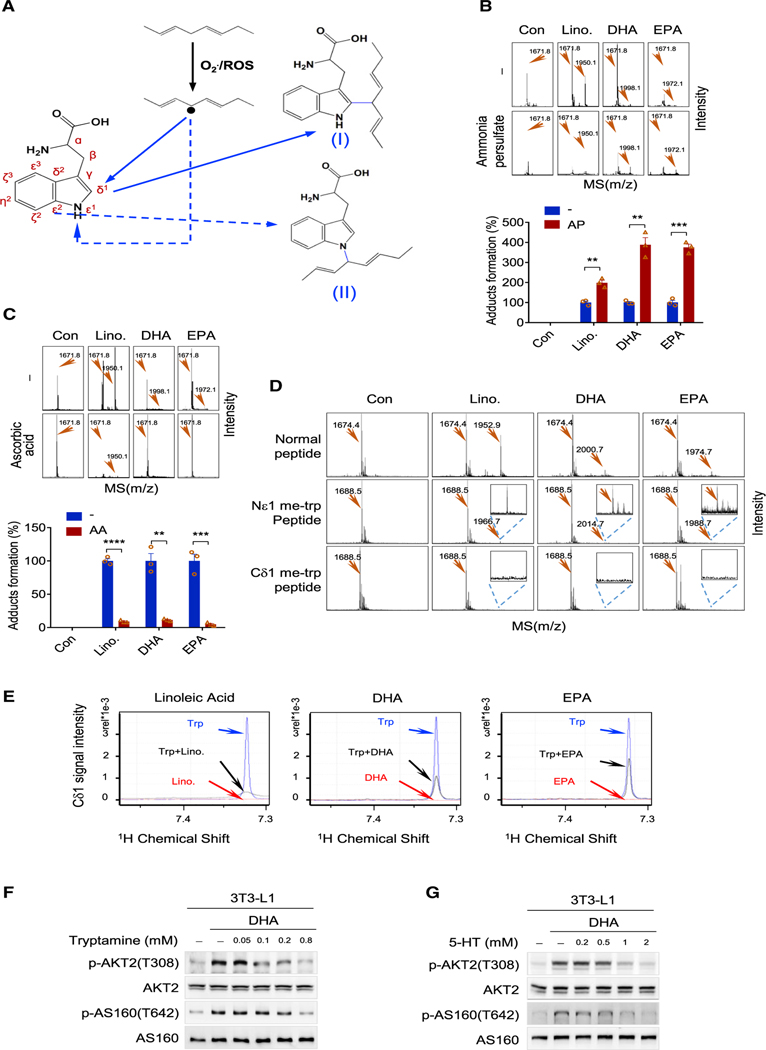
Figure 3. Cδ1 of tryptophan is the site of methylene bridge acylation.
(A) Proposed mechanisms for methylene bridge-tryptophan adduct formation. After induction by other free radicals, methylene bridge free radicals may attack either Cδ1 and Nɛ1 of tryptophan to form either adduct I or II, respectively.
(B) Free radical inducer promotes tryptophan acylation. The formation of EPA, DHA, and LA adducts in a peptide with or without ammonia persulfate in thereaction mix was analyzed by MS (upper), and adducts levels were measured (lower) (n = 3, mean ± SEM).
(C) Free radical scavenger inhibits tryptophan acylation. The formation of the EPA, DHA, and LA adduct in a peptide with or without ascorbic acid in the reaction mix was analyzed by MS (upper), and adducts levels were measured (lower) (n = 3, mean ± SEM).
(D) Cδ1 of tryptophan is the acylation site by a methylene bridge. A tryptophan-containing peptide (Ac-FTEGAFKDWGYQLA) and peptides with the same sequence but with tryptophan replaced by Nɛ1 or Cδ1 methyl-blocked tryptophan were tested for their abilities to be adducted by linoleic acid, EPA, and DHA. Adduct formation was assayed by MS.
(E) NMR analysis of the reactivities of Cδ1 of tryptophan. The NMR signal of Cδ1 of tryptophan was decreased by LA (left), DHA (middle), and EPA (right).
(F and G) AKT signaling activation by DHA was reversed by tryptamine (F) and 5-hydroxy-tryptamine (5-HT) (G). The levels of p-AKT2(T308) and p-AS160(T642) were determined in 3T3-L1 cells cultured in media supplemented with DHA, DHA/tryptamine, or DHA/5-HT.