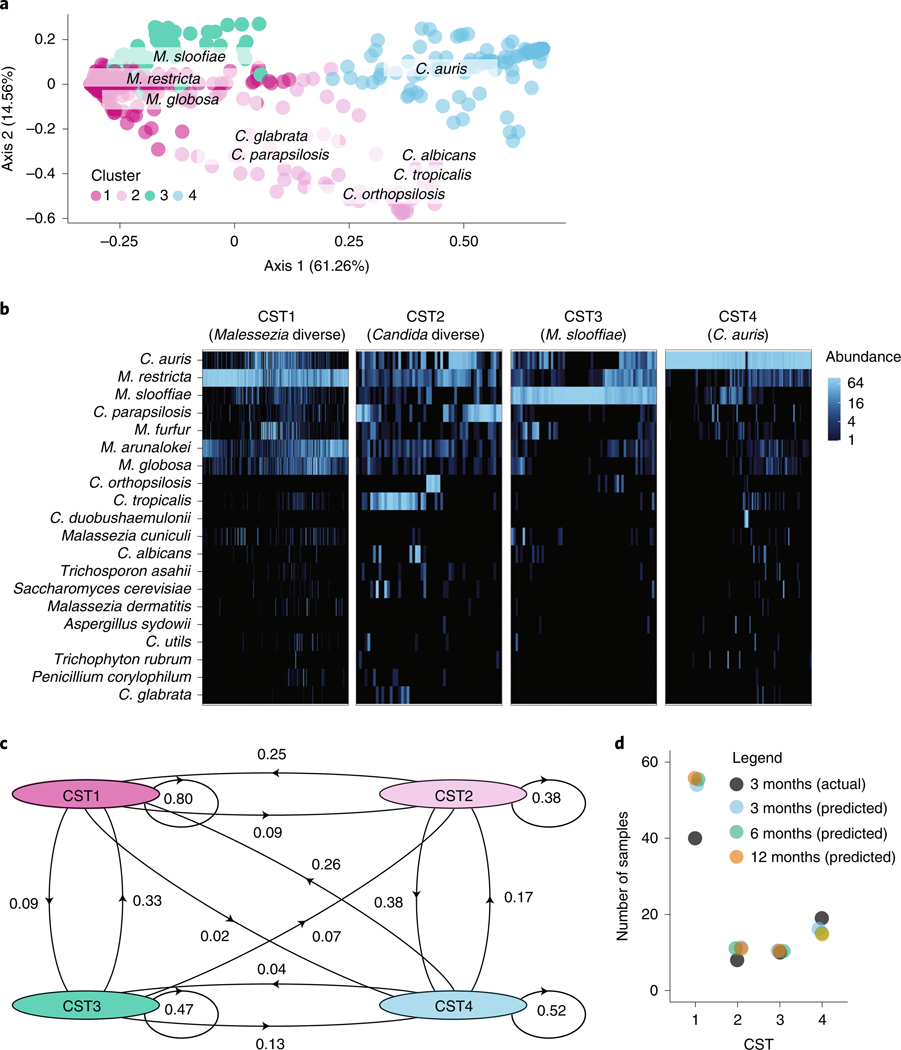
Fig. 4 |. Skin fungal communities, dominated by Malassezia and Candida species, have differential stability, resilience and likelihood of invasion by C. auris.
a, Principal-coordinate analysis of the weighted UniFrac metric of the fungal community at each body site (toe webs, fingertips and/or palm, inguinal crease, anterior nares). Samples are shaded according to CST identity, as revealed by ‘partition around medoids’ analysis. CST1 tends to be dominated by M. restricta (N = 256, 53.0%), CST2 by a variety of Candida species (N = 52, 10.7%), CST3 by M. slooffiae (N = 62, 12.8%) and CST4 by C. auris (N = 113, 23.4%). Segregation of Malassezia and Candida species across the first axis explains 61% of the variance. Candida species segregate across the second major axis, which accounts for ∼15% of the variance. b, Relative abundance of the top 20 species in each sample, clustered by CST. Shading is based on the relative abundance of taxa within each sample. c, Self and interstate transition probabilities inferred for samples of the toe webs, palm and/or fingertips, inguinal crease and anterior nares. d, Scatterplot of the predicted numbers of samples in each CST at 3, 6 and 12 months after sample collection compared to the actual number of samples at 3 months. Predictions were generated using the Markov chain in c.