Figure 1.
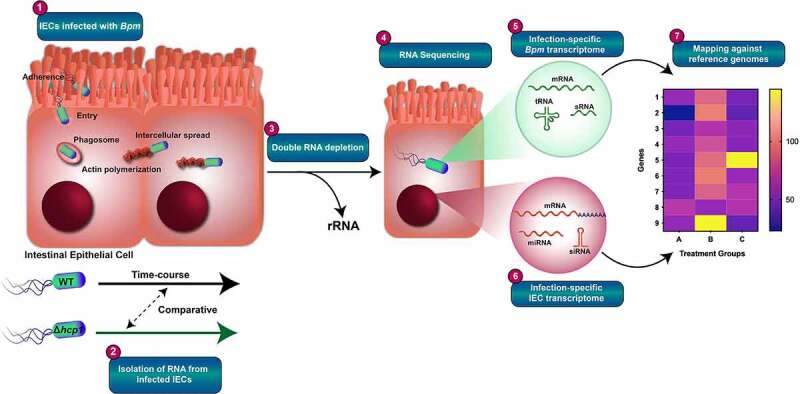
Workflow of dual RNA-seq experiment. High-quality RNA was collected from primary intestinal epithelial cells (IECs) and enriched for either host or bacteria RNA transcripts before creating cDNA libraries. 1) The IECs were infected with Bpm WT K96243 and a Bpm Δhcp1 mutant for 6 and 12 h using an MOI of 10. 2) Total RNA was isolated from infected cells and purified RNA samples used for downstream analysis. 3) Bacterial or host rRNA was depleted using an RNA purification kit. 4) Purified RNA samples were sequenced using Illumina technology. 5) Host RNA libraries were mapped against a parent genomic sequence. 6) Bacterial RNA libraries were mapped against a parent Bpm sequence. 7) Differences in expression profiles were presented as heatmap and volcano plots.
