Figure 6.
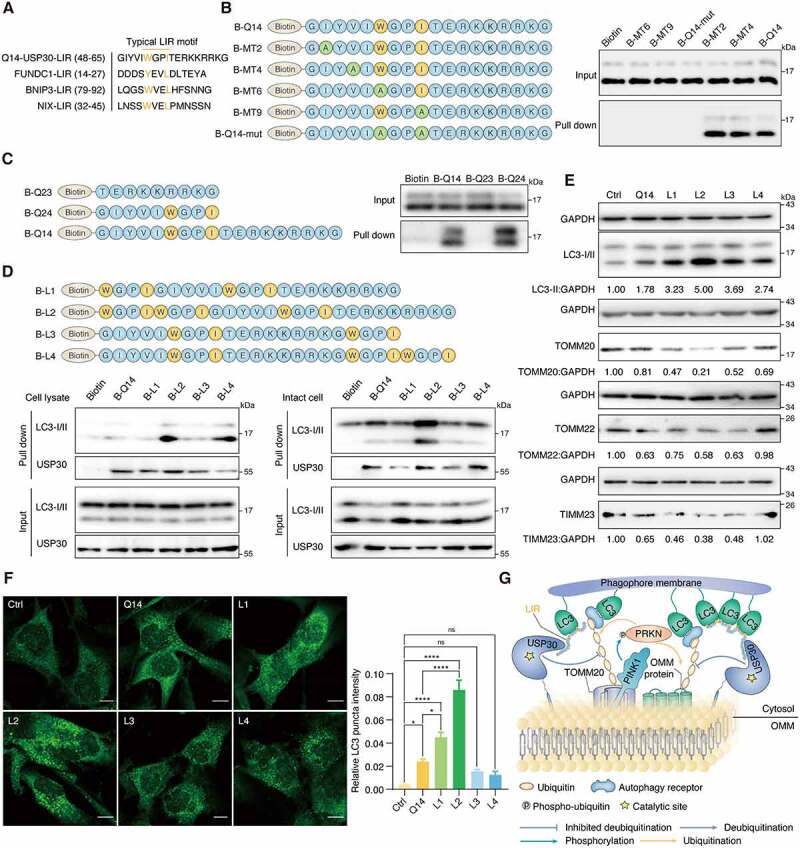
Q14 peptide contains the LIR and interacts with LC3B in A172 cells. (A) Typical LIR motifs of mitophagy related proteins were identified from iLIR database and aligned manually alongside USP30 for comparison. The colored residues indicate highly conserved residues in the putative LIR of USP30. (B) A172 cell lysates were incubated with biotin, single mutant peptide MT2 (I/A), MT4 (V/A), MT6 (W/A) and MT9 (I/A), double mutant peptide Q14-mut (WxxI/AxxA) and biotin-labeled Q14 peptide (B-Q14) at 4°C. SA beads were added into the mixture and used for Western blot analysis through an anti-LC3B antibody. Mutations only at the LIR motif (MT6, MT9, Q14-mut) but not at other motifs (MT2, MT4), significantly weakened their interactions with LC3. The peptide sequences are shown on the left. (C) Peptides contain with LIR motif (B-Q14, B-Q24) could pull down LC3 while the other segment (B-Q23) could not. The peptide sequences are shown on the left. (D) The lysate and intact-cell based pull down assay performed by B-Q14 peptide and peptides with extended LIR motifs (B-L1, B-L2, B-L3, B-L4). The peptide sequences are shown on the top panels. (E) Immunoblotting for LC3-II, TOMM20, TOMM22 and TIMM23 was assessed in A172 cells incubated with peptide (Q14, L1, L2, L3, L4, 50 µM) for 24 h. GAPDH was used as a loading control. The relative band intensities are quantified by image-J software and shown below the blots. (F) Immunofluorescence showing the endogenous LC3 puncta in A172 cells incubated with DMSO, Q14, L1, L2, L3 and L4 peptide. Scale bar: 10 μm. Quantification of the LC3 puncta intensities in the cells. Data are presented as mean ± s.e.m. from at least 300 cells. Data were analyzed by one-way ANOVA (multiple comparisons) using GraphPad Prism 8.0. * P< 0.05, ****p < 0.0001. (G) Q14 peptide binds with USP30 and inhibits its deubiquitinating activity. Besides, through interacting with LC3 by its LIR motif, Q14 peptide links the phagophore membrane to USP30 and ubiquitinated mitochondria, promoting the mitophagy level. Experiments in B, C, D and E were repeated three times independently with similar results. Full-length gels for B, C, D and E are in Fig. S8.
