Figure 2.
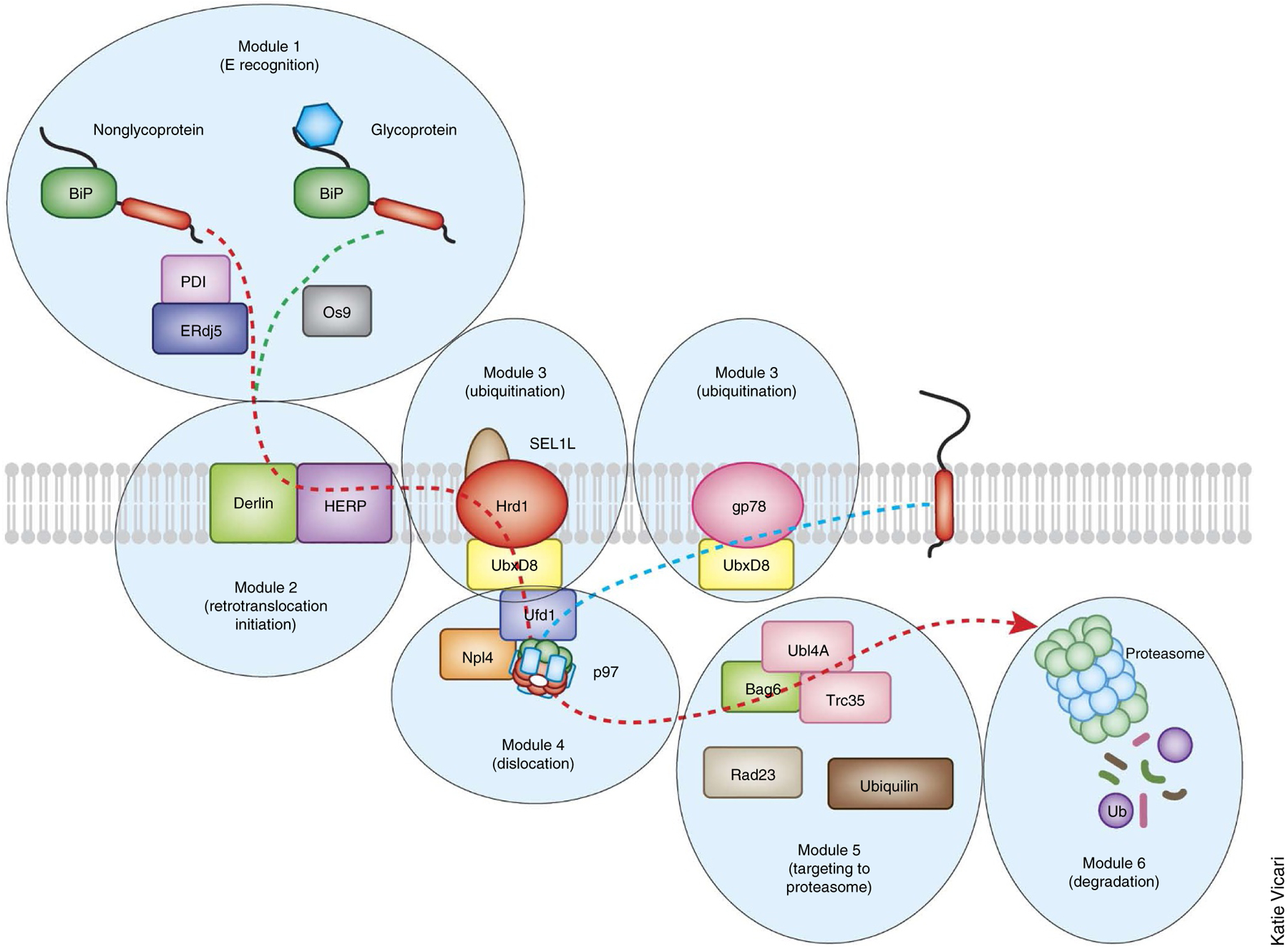
Modular organization of the mammalian ERAD system. A large number of ERAD factors are organized into several modules to mediate retrotranslocation. In general, proteins in the same module tend to form stable interactions, whereas factors from different modules bind each other in a dynamic manner, although there are exceptions. For instance, a large, stable membrane complex consisting of an E3 ligase and other factors is likely to mediate the retrotranslocation initiation and the ubiquitination modules. Within a given module, factors can also form parallel pathways to transport distinct classes of the substrates. For an ERAD-L mechanism, the module for substrate recognition and recruitment distinguishes misfolded proteins from folded ones in the lumen and then selectively targets them for insertion into the ER membranes. Subsequent ubiquitin conjugation is mediated by ubiquitination modules. This is followed by extraction from the ER membrane via the AAA+ ATPase p97/VCP complex. Dislocated products are shuttled to the proteasome, which may have been brought near to the retrotranslocation site through interaction with the ER membranes. The yeast ERAD system has a similar modular organization with fewer participating components. The retrotranslocation and degradation of ERAD-M and ERAD-C substrates do not seem to require the recognition and initiation modules in the ER. The dashed lines indicate the flow of the substrates.
