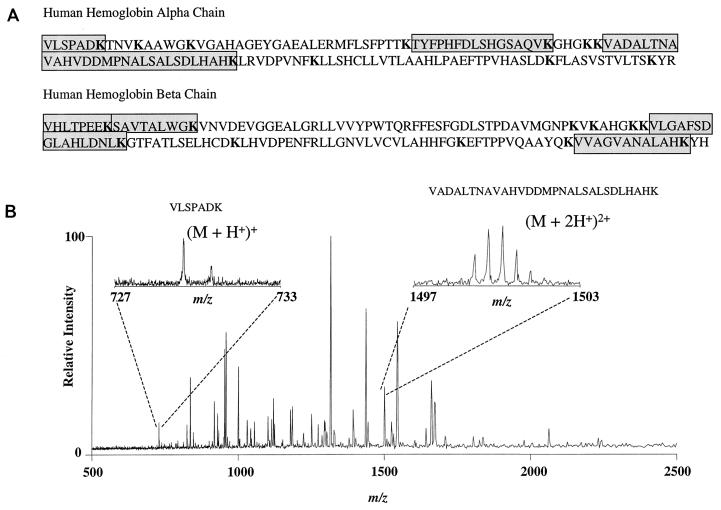
FIG. 7.
Mass spectrometric analysis of Kgp-digested hemoglobin fragments. (A) Amino acid sequence of human α and β hemoglobin chains. Lysine residues are indicated in boldface; amino acid sequences of peptides demonstrated by ESI-FTICR spectroscopy are shaded. (B) ESI-FTICR mass spectrum of a human hemoglobin sample digested by the lysine-specific cysteine protease Kgp. The hemoglobinolytic potential of Kgp is verified by the observance of expected hemoglobin digestion fragments. The resolved carbon-13 isotopic distribution is evident in the two insets for two of the hemoglobin fragments selected as examples. Additional peaks in the spectrum not related to hemoglobin are suggestive of autodigestion of the protease.