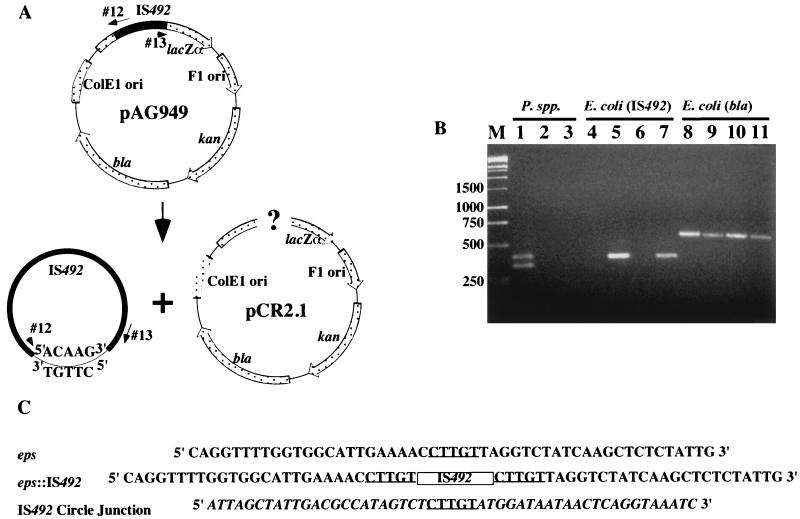
FIG. 2.
Detection of IS492 circles in vivo by PCR analysis. (A) Diagram of IS492-containing plasmid pAG949 and locations of primers (oligonucleotides 12 and 13) used in this PCR analysis. Potential products of excision are drawn below the diagram. (B) Agarose gel electrophoresis of products generated from colony PCR with the circle junction primers (oligonucleotides 12 and 13) and total DNA from P. atlantica DB27 (lane 1), P. espejiana (lane 2), P. haloplanktis (lane 3), E. coli InvαF′ containing pCR2.1 (lane 4), InvαF′ containing pAG949 (lane 5), E. coli HMS174(DE3) containing pCR2.1 (lane 6), or HMS174(DE3) containing pAG949 (lane 7). Lanes 8 to 11 have the same template DNAs as lanes 4 to 7, respectively, but PCR was performed with the bla primers (see Materials and Methods), which amplify the β-lactamase gene of pAG949, as a control for template addition. Lane M, molecular size markers in base pairs. (C) Sequences of the eps locus prior to and after IS492 insertion and the circle junction sequence with flanking IS492 termini. The 5-bp target duplication-circle junction sequence is underlined, the IS492 insertion in eps is represented by a box, and the IS492 sequence is italicized in the circle junction sequence.