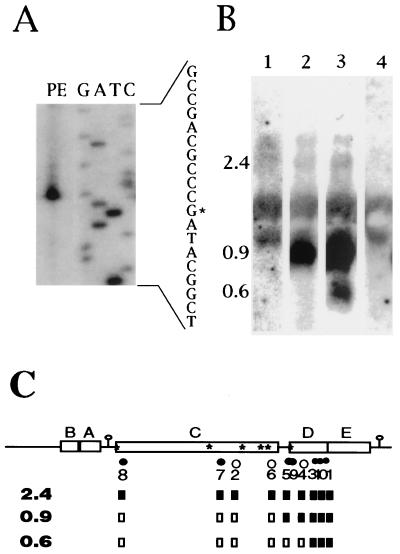
FIG. 2.
PE and RNA blot analyses of the puc operon using oligonucleotides as primers or probes. (A) Results of a PE experiment that utilized the ALPE3 primer and RNA from SB1003. The 5′ end at an RNA G residue corresponds to the proposed start site of a promoter located within pucD. (B) Results of an RNA blot experiment that utilized oligonucleotide probes ALPE6 (lane 1), ALPE4 (lane 2), and ALPE1 (lane 3, 4) hybridized to RNA from ΔLHII(pBACDE) (lanes 1 to 3) or ΔLHII(pRK415) (lane 4). RNA from a strain lacking the puc operon (lane 4) shows only nonspecific hybridization to rRNA. (C) Summary of PE and RNA blot experiments. The locations of sequences complementary to oligonucleotides are indicated by dots and ALPE numerical designations below a schematic of the puc operon. Open dots indicate that no ends were detected (within 60 nucleotides) by PE using that primer, whereas filled dots show that an end was detected. These oligonucleotides were also used to probe RNA blots. If a given primer detected the 2.4-, 0.9-, or 0.6-kb transcript, the corresponding box below the primer number is filled whereas open boxes represent negative results. Inverted repeats are shown as loops with stems, and the positions of the six RNase E consensus sites are shown (∗).