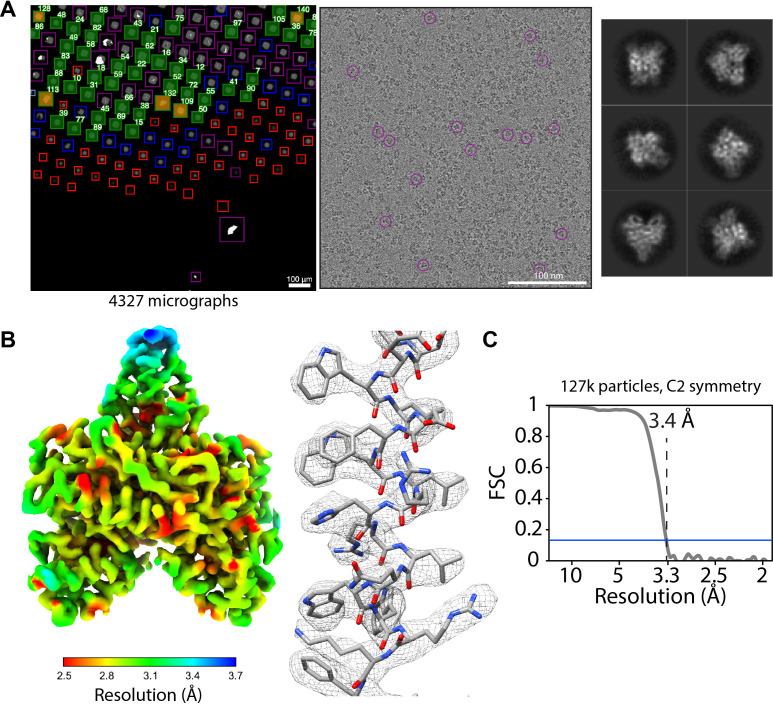
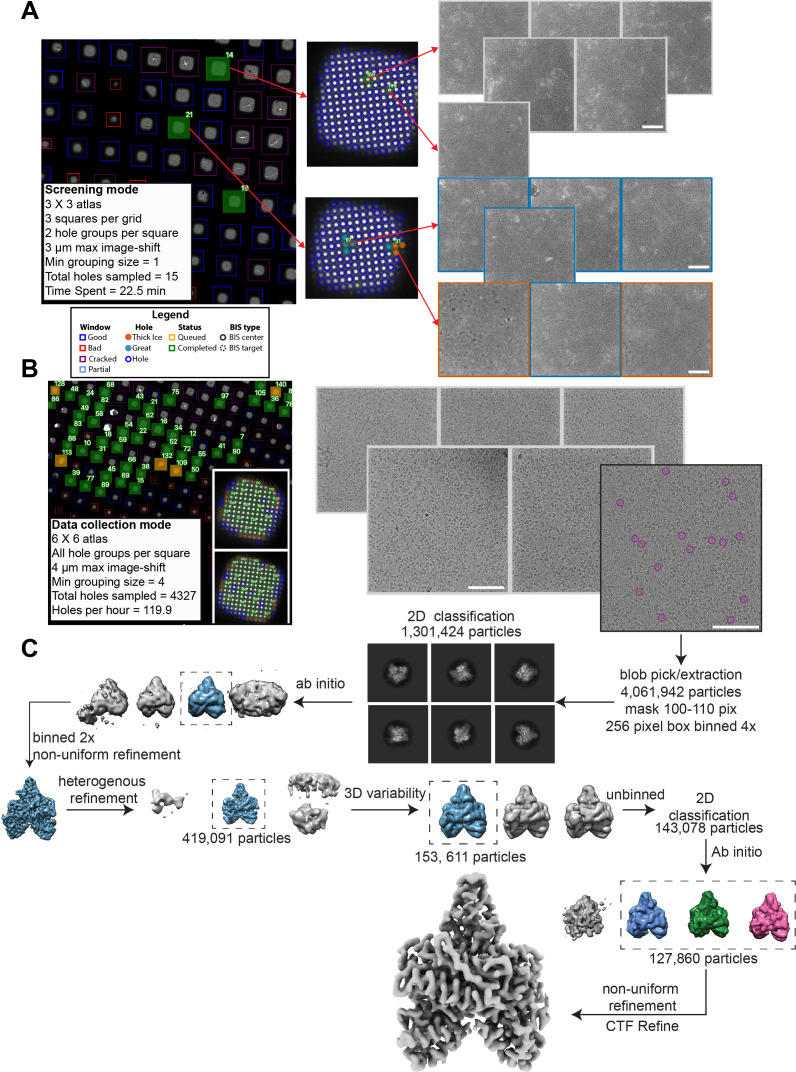
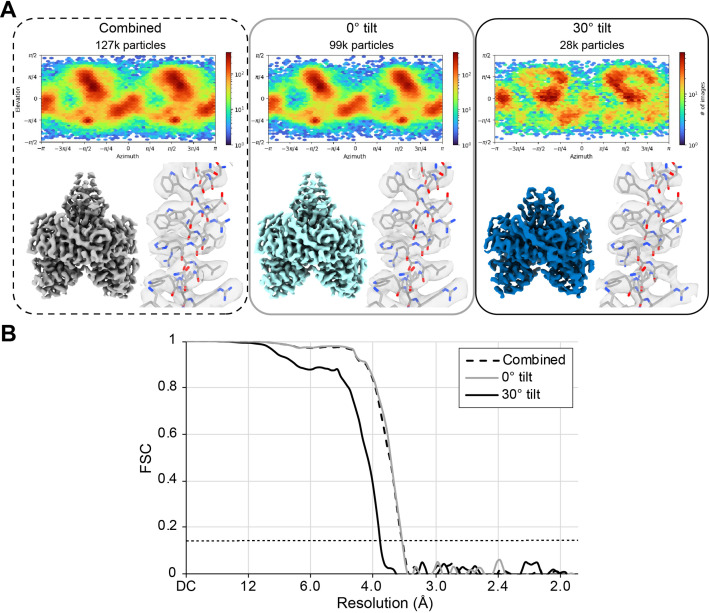
Figure 4. Acquisition of POLG2 dataset using SmartScope.
(A) Atlas of the specimen (left), typical micrograph (center) with some particles picked (purple circles) and 2D classes of POLG2 (right). (B) Resulting map of POLG2 colored by local resolution (left) and example of an alpha helix with atomic model fit into the density (left). (C) Masked Fourier-shell correlation curve between half-maps showing a resolution of 3.4Å.