Figure 3.
Immunostimulatory RNAs induce IFN-I production through RIG-I-IRF3 pathway
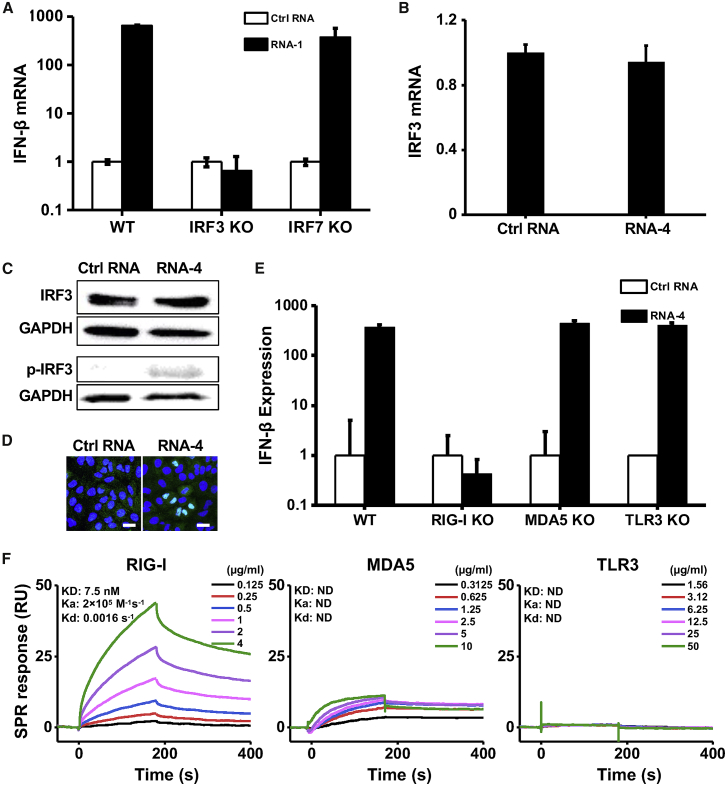
(A) IFN-β mRNA levels in wild-type (WT) HAP1 cells, IRF3 knockout HAP1 cells, or IRF7 knockout HAP1 cells after transfection with RNA-1 or scrambled RNA control for 48 h. Data are shown as fold change relative to the scrambled RNA control (N = 3). Note that IRF3 knockdown completely abolished the IFN-β response. (B) IRF3 mRNA levels measured in A549 cells transfected with immunostimulatory RNA-4 or a scrambled RNA control (data are shown as fold change relative to the control RNA; N = 3). (C) Total and phosphorylated IRF3 protein levels in A549 cells at 48 h after transfection with RNA-4 or scrambled RNA control. GAPDH was used as a loading control. (D) Immunofluorescence micrographs showing the distribution of phosphorylated IRF3 in A549 cells transfected with RNA-4 or scrambled RNA control at 48 h post transfection (green, phosphorylated IRF3; blue, DAPI-stained nuclei; arrowheads, nuclei expressing phosphorylated IRF3). Scale bar, 20 μm. (E) IFN-β expression in WT A549-Dual cells, RIG-I knockout A549-Dual cells, MDA5 knockout A549-Dual cells, or TLR3 knockout A549 cells at 48 h after transfection with immunostimulatory RNA-4 or a scrambled RNA control. Data are shown as fold change relative to the scrambled RNA control; N = 6. Note that RIG-I knockout abolished the ability of the immunostimulatory RNAs to induce IFN-β. (F) SPR characterization of the binding affinity between cellular RNA sensors (RIG-I, MDA5, and TLR3) and RNA-1. Equilibrium dissociation constant (KD), association rate constant (Ka), and dissociation rate constant (Kd) are labeled on the graphs. (A, B, and E) Data are shown as means ± SD.