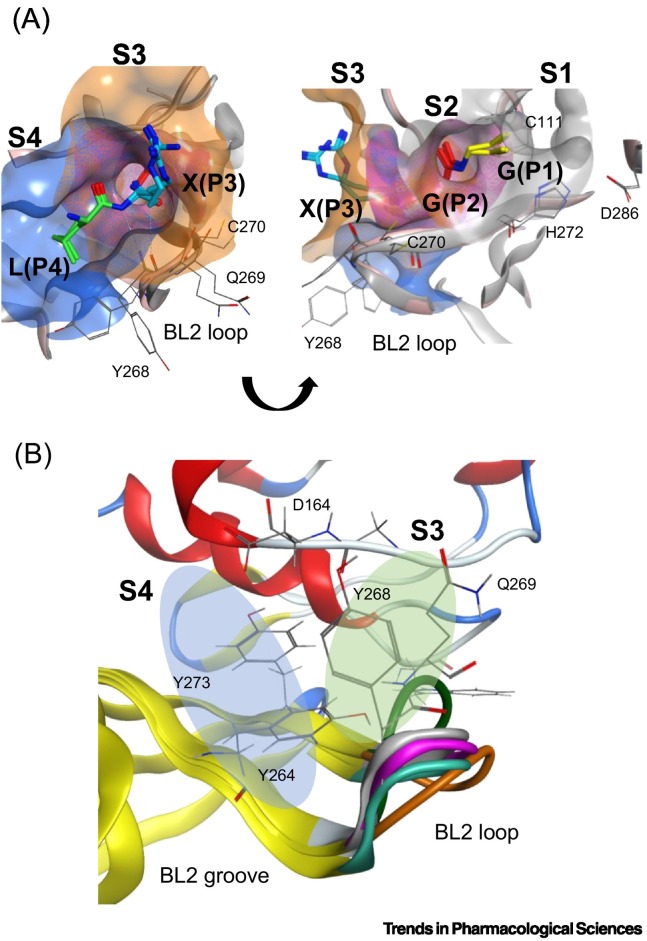
Figure 2.
The active site of SARS-CoV-2 PLpro.
(A) PLpro's active site recognizes a ‘LXGG’ consensus sequence on viral polyproteins, ISG15, and ubiquitin. The PLpro is characterized by a Gly-Gly tunnel in the S1 and S2 subsites, and more accommodating S3 and S4 subsites. The active site is delimited into four subsites, each occupied by a residue of the substrate sequence. Residues of ISG15 and ubiquitin substrates are colored per the occupied position in the active site: green: P4 Leu; teal: P3 Arg; red: P2 Gly; yellow: P1 Gly. The surface of the subsites is colored per the substrate residues, in order from S4 (blue), S3 (orange), S2 (magenta), and S1 (gray). Important PLpro active site residues are shown in thin gray sticks and labeled appropriately. PDB used: 7RBR for ubiquitin and 7RBS for ISG15. Pink ribbons: PLpro with ISG15; gray ribbons: PLpro with ubiquitin. (B) The BL2 loop, spanning residues 267-271 in the S3 and S4 subsites, is highly flexible. The BL2 loop undergoes induced-fitted effects and enables the active site to accommodate different binding partners by transitioning between open and closed states. S4 subsite shown in the blue oval and S3 subsite shown in the green oval. The ‘BL2 groove’ is adjacent to the S4 and is formed when the BL2 loop is in its closed conformation. Orange: apo (PDB ID: 6W9C); magenta: Ub (PDB ID: 6XAA); teal: VIR250 (PDB ID: 6WUU); white: ISG15 (PDB ID: 6XA9); green: GRL-0617 (PDB: 7CMD).