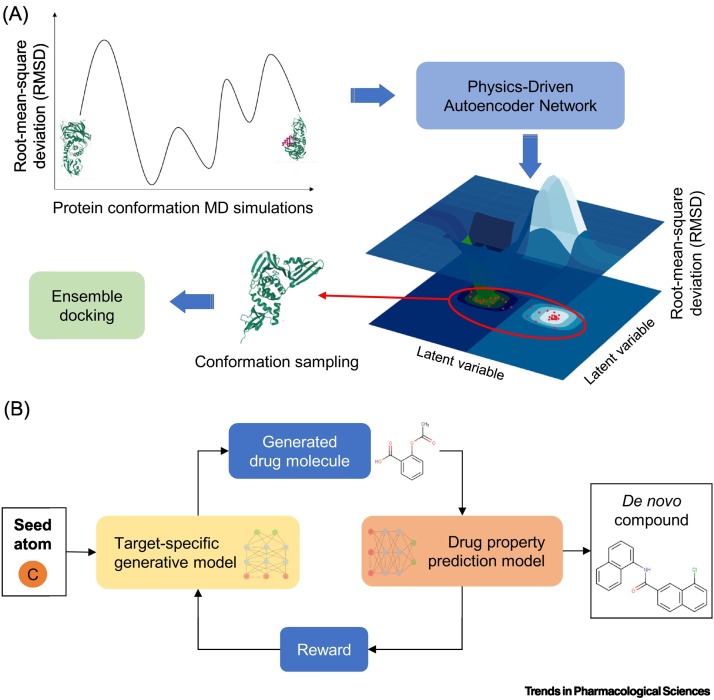
Figure 3.
Machine-learning-assisted PLpro drug discovery.
(A) Physics-based neural networks can learn the protein conformational space. The method returns new, plausible near ligand-bound protein structures from the protein conformational space, complementing the existing ones. Specifically, molecular dynamics (MD) simulations generate many protein structures, encompassing all the possible conformations that a protein can adopt, from unbound to bound. A sample of the structures obtained from the MD simulations are used to train a generative autoencoder that correctly reconstructs protein conformations to account for broad flexibility upon ligand binding. High-quality structures (red dots on the 3D latent space) outputted from the autoencoder are selected for SBVS, such as ensemble docking. Being able to incorporate different conformations will enable SBVS to capture virtual hits that would otherwise be missed due to not having the correct crystal structure. (B) Pipeline of a modern deep reinforcement learning (DRL)-based methodology for de novo compound generation used to enhance new drug design. A generative model takes in a seed atom to start the process and continually predicts subsequent atoms of a compound (in SMILES format) until the model produces a valid chemical structure. The ‘drug property prediction model’ checks the biological, chemical, and physical properties of the newly generated molecules, and the model calculates a reward based on the selected features. Depending on the reward threshold, the generative model either further modifies the molecule or stops the generative process. The exploration of this space is important to generate compounds designed for the target of interest. Abbreviation: SBVS, structure-based virtual screening.