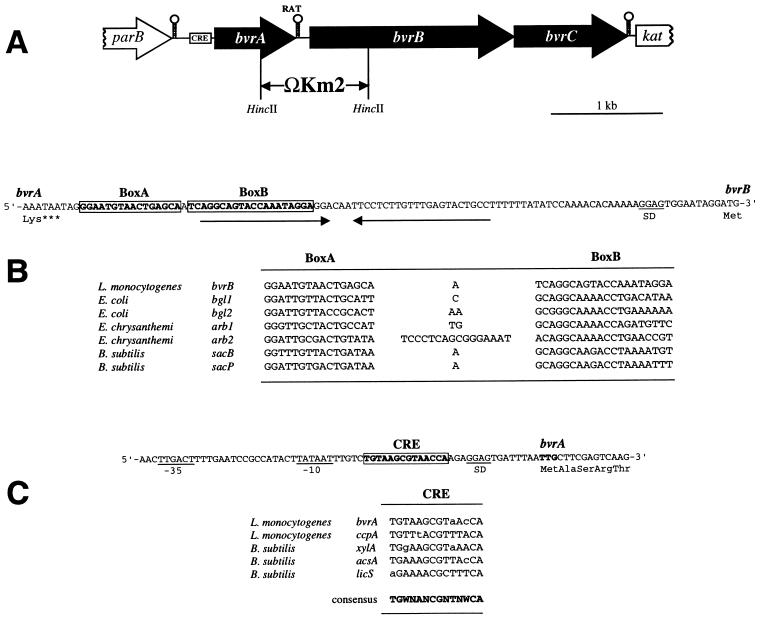
FIG. 2.
(A) Scheme of the genetic structure of the bvr locus of L. monocytogenes. The positions of the putative transcriptional terminators, CRE, and RAT sequences are shown. The bvr mutation was created by allele exchange by using a plasmid construct in which a HincII fragment encompassing the 3′-terminal third of bvrA and the 5′-proximal third of bvrB was replaced by the ΩKm2 interposon, as indicated. (B) Nucleotide sequence of the intergenic region between bvrA and bvrB (the respective last and first codons are shown) and position of the putative RAT sequence (boxed), which overlaps the 5′ part of a palindromic structure that may act as transcriptional terminator (indicated by inverted arrows). Below, comparison of the putative RAT sequence preceding bvrB with known RAT sequences (13). (C) Nucleotide sequence of the 3′ region upstream from bvrA (its TTG start codon is in boldface type) showing −10 and −35 putative promoter sequences and the CRE-like sequence (boxed). Below, comparison of the putative CRE preceding bvrA with the CRE-like element of the ccpA promoter region of L. monocytogenes (3) and known active CRE sites from gram-positive bacteria (21). Deviations from the CRE consensus sequence (in boldface) are shown in lowercase (N = any nucleotide; W = A or T).