Figure 1.
Workflow overview
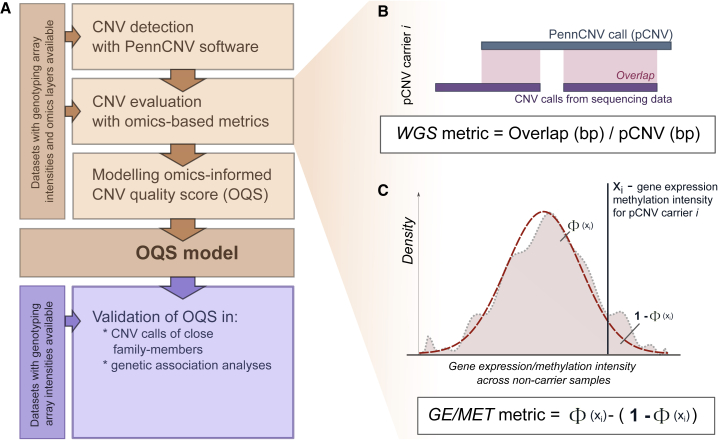
(A) Quality estimation and modeling pipeline for PennCNV copy-number variation calls (pCNVs).
(B and C) The pCNV quality metrics are estimated based on (B) whole-genome sequencing (WGS) data and (C) gene expression (GE) and/or overall methylation (MET) intensity of genes/CpG sites overlapping the corresponding CNV calls.
(B) WGS metric is a fraction of pCNV that can be mapped to WGS CNVs of the same individual.
(C) To calculate GE/MET metrics, the reference distribution of expression/intensity based on non-carriers (pink area) is approximated to standard normal distribution (red dashed line), and the Z score of the expression/intensity of each pCNV carrier (xi) is compared with it one at a time. The metric is a difference between the fraction of non-carriers with the corresponding value ≤xi and those with the corresponding value >xi and captures how extreme xi is compared with the reference distribution of non-carriers. In case a pCNV overlaps with several genes/CpG sites, the metric values are averaged over them.