Figure 4.
Impact of OQS on CNV-trait associations
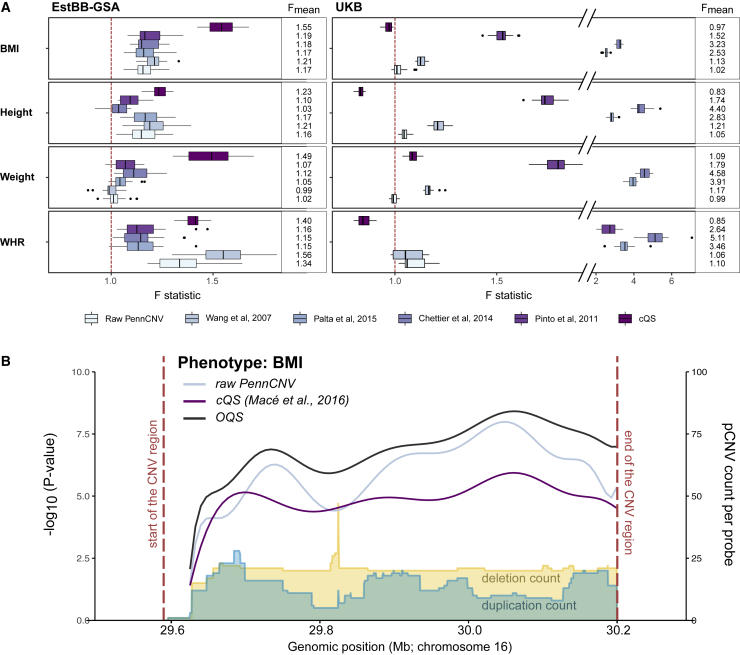
(A) Change of variance explained in mirror-type model when using OQS over raw PennCNV, four published quality filtering approaches,18,20,38,39 or cQS19 in the EstBB-GSA and UKB, depicted as distribution of F statistics calculated by randomizing the probe pruning priority order 20 times (see material and methods). Explained variance is increased when F >1 and decreased when F <1. Larger F values indicate greater improvement in statistical power when using OQS over the given reference approach.
(B) Locus plot of a CNV region in 16p11.2 BP4-BP5 (red dashed lines: chr16:29,590,000–30,200,000 in GRCh37) associated with BMI in EstBB-GSA dataset. The lines indicate the –log10 association p values using mirror model with raw PennCNV calls (light blue), cQS (purple), and OQS (black). The yellow and blue areas illustrate the frequency of PennCNV deletion and duplication counts, respectively, across the region.