Figure 1.
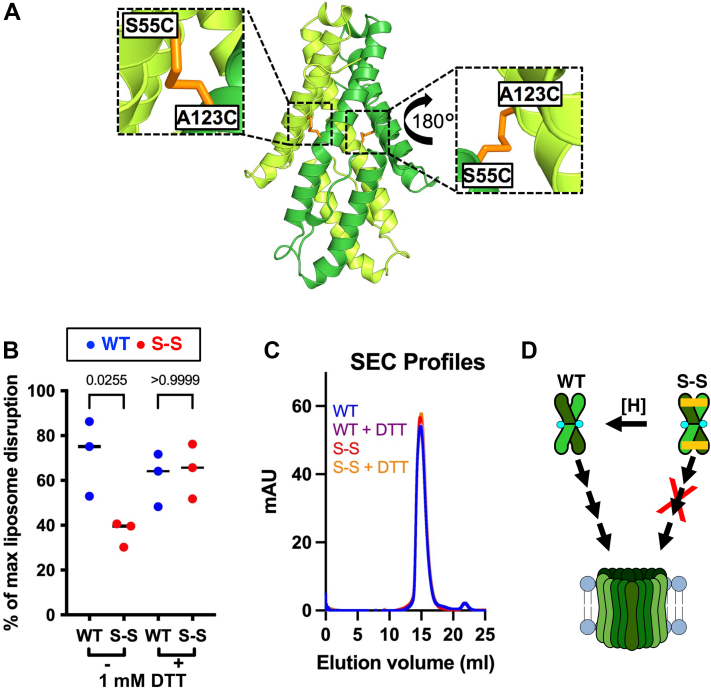
Disulfide-locked CelTOS dimer is inactive in pore formation.A, CelTOS dimer (PDB ID: 5TSZ) is shown in the cartoon highlighting the location of Ala123 and Ser55. Note that the shown modeled structure highlights residues that were computationally mutated to cysteines to lock CelTOS in the dimer state. B, pore-forming assay: disulfide-locked CelTOS dimer (S–S) is significantly less active than the WT protein. Note that this loss of activity was rescued in the presence of 1 mM DTT that reduces the disulfide bridge, enables dimer dissociation, and facilitates pore formation. The graph represents the median value and three independent biological replicates each with three to four technical replicates (please note that the graph shows the combined data from three individual biological replicates demonstrating the reproducibility of the data. Please see Fig. S3 for these three individual biological replicates). Significance was determined using Kruskal–Wallis analysis and Dunn’s multiple comparison. C, size-exclusion chromatography (SEC) profiles of WT and S–S in the presence or the absence of 1 mM DTT. D, a model depicting the plausible mechanism of conversion of soluble CelTOS dimer to the multimeric membrane pore (left); S–S inactivation (right) and its rescue by disulfide reduction (shown as [H]). CelTOS, Cell-traversal protein for ookinetes and sporozoites; PDB, Protein Data Bank.