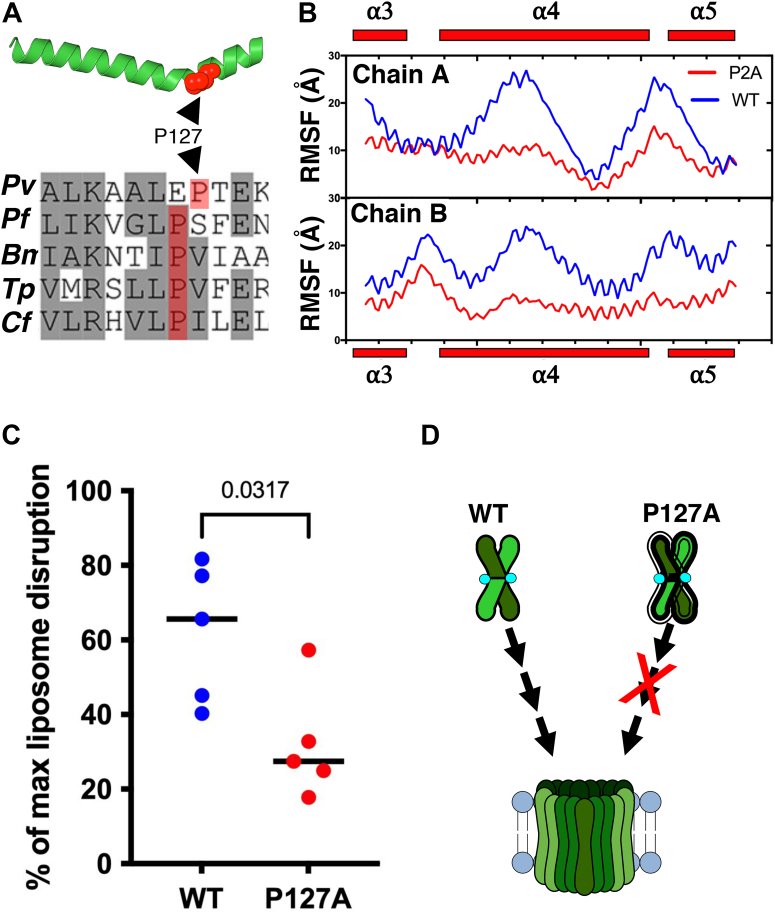
Figure 2.
Pro127 is required for pore-forming activity of CelTOS.A, CelTOS helix 4 (PDB ID: 5TSZ) is shown. Pro127 (shown in red sphere) causes a bend in the helix 4 (top panel). Multiple sequence alignment (MSA) of CelTOS orthologs in apicomplexan parasites showing conserved Pro127 (highlighted in red). Residues in gray background represent conserved residues, and residues in white background represent nonconserved residues. Bm, Babesia microti; Cf, Cytauxzoon felis; Pf, Plasmodium falciparum; Pv, Plasmodium vivax; Tp, Theileria parva (bottom panel). B, root mean square fluctuation (RMSF) plot of two monomers indicates that the Pro127Ala (P127A) mutant is less flexible in different regions including helix 4 (⍺4). C, pore-formation assay of CelTOS-WT and Pro127Ala (P127A) mutant. The graph represents the median value and five independent biological replicates each with eight technical replicates (see Fig. S6 for five independent biological replicates demonstrating the reproducibility of the data). Significance was determined using a Mann–Whitney U test. D, a model showing the enhanced rigidity in P127A mutant causes loss of pore-forming activity. CelTOS, Cell-traversal protein for ookinetes and sporozoites; PDB, Protein Data Bank.