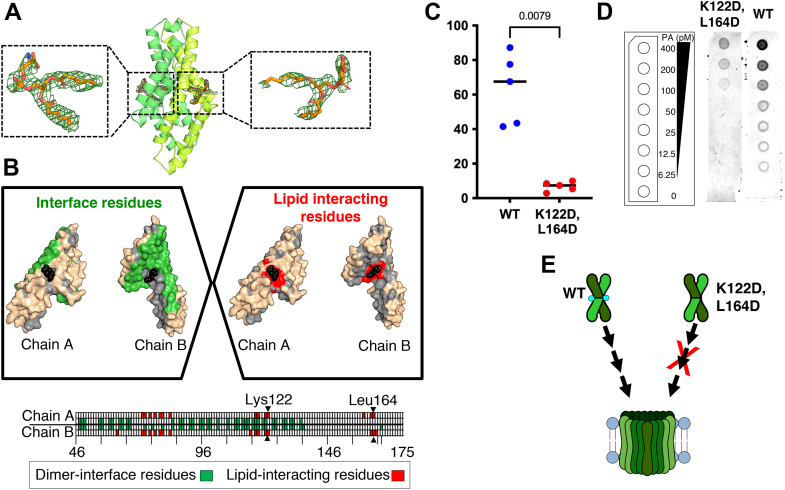
Figure 4.
Lys122 and Leu164 are important for pore-forming activity of CelTOS.A, cartoon view of CelTOS structure (PDB ID: 5TSZ). The two monomers are shown in green and lime colors. Two POPA molecules (blue sticks) modeled in the electron densities are shown in the enlarged views. 2fo–fc map is contoured at 1.0 sigma. B, top, the two monomer structures of CelTOS showing the interface residues in green (left). CelTOS monomers showing the plausible lipid-interacting residues in red (right). Bottom, an illustration of the CelTOS dimer showing the residues that are involved at the monomer–monomer interactions (green in color). Residues within 5 Å radius of bound lipid molecules are highlighted in red. Lys122 and Leu164 are shown. C, pore-formation assay of Lys122Asp,Leu164Asp(K122D,L164D) mutant and its comparison to the WT protein. The graph represents the median value and five independent biological replicates each with eight technical replicates (see Fig. S9 for five independent biological replicates demonstrating the reproducibility of the data). Significance was determined using a Mann–Whitney U test. D, evaluation of binding to phosphatidic acid of WT and K122D,L164D mutant. An equimolar protein concentration of 1 μM was used for WT and K122D,L164D mutant. Left, a schematic layout of the phosphatidic acid (PA) lipid strips containing a concentration gradient of PA from 400 to 0 pmol. Middle, the double mutant shows poor binding to PA. Right, the WT protein shows binding to PA. One representative biological replicate of three is shown. Two additional biological replicates are shown in Fig. S10. E, a model showing the loss of pore-forming activity of the double mutant (K122D,L164D). CelTOS, Cell-traversal protein for ookinetes and sporozoites; PDB, Protein Data Bank; POPA, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate.