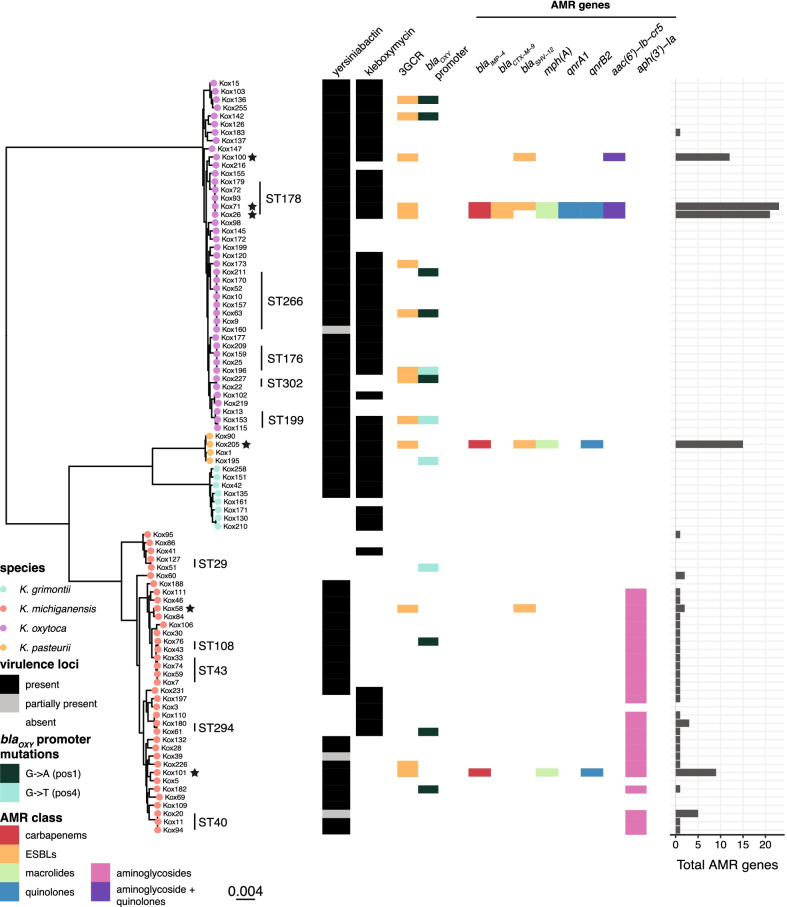
Fig. 1.
Core genome phylogeny of the KoSC. Left, maximum-likelihood core genome phylogeny (midpoint rooted) of all 92 sequenced isolates in this study, with tips coloured by species. Scale indicates substitutions per site. STs with ≥ 2 members are labelled. Black stars indicate isolates with completed genomes. Heatmap shows presence (coloured) or absence (white) of the virulence loci kleboxymycin and yersiniabactin (as per legend), in addition to resistance to third-generation cephalosporins, key AMR mutations and genes (coloured by mutation type or resistance gene class as per legend). Barchart indicates total number of acquired AMR genes per isolate