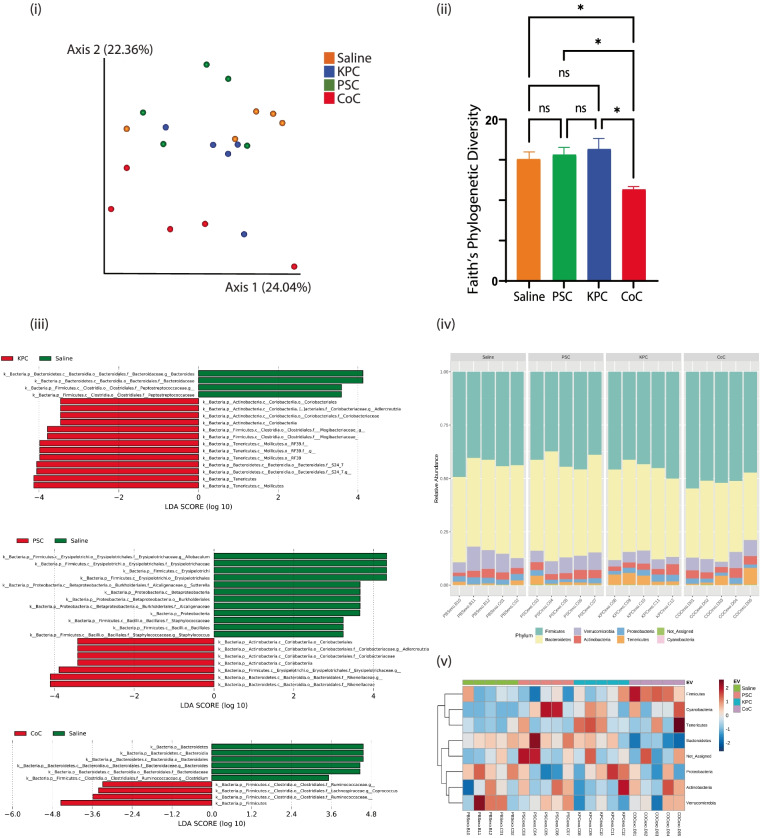
Fig. 4.
(i) Principal coordinate analysis (PCA) plot of weighted UniFrac distances (metric of β diversity) with q < 0.01 among all four groups (ii) Faith’s phylogenetic diversity (metric of α diversity) at sequencing depth of 19,200; * represents p < 0.05; ns represents not significant (iii) LefSe (Linear Discriminant Analysis Effect Size) analysis of bacterial samples among samples KPC Vs PBS control, PSC Vs PBS Control, and CoC (Co-culture) Vs PBS control. The top discriminative bacterial taxa identified between the represented conditions is shown. (iv) Phylum level bacterial composition in the different experimental conditions. Samples include PBS, KPC, CoC and PSC conditions. N = 5 samples were considered for the analysis. (v) Heatmap of OTU abundance at the phylum level