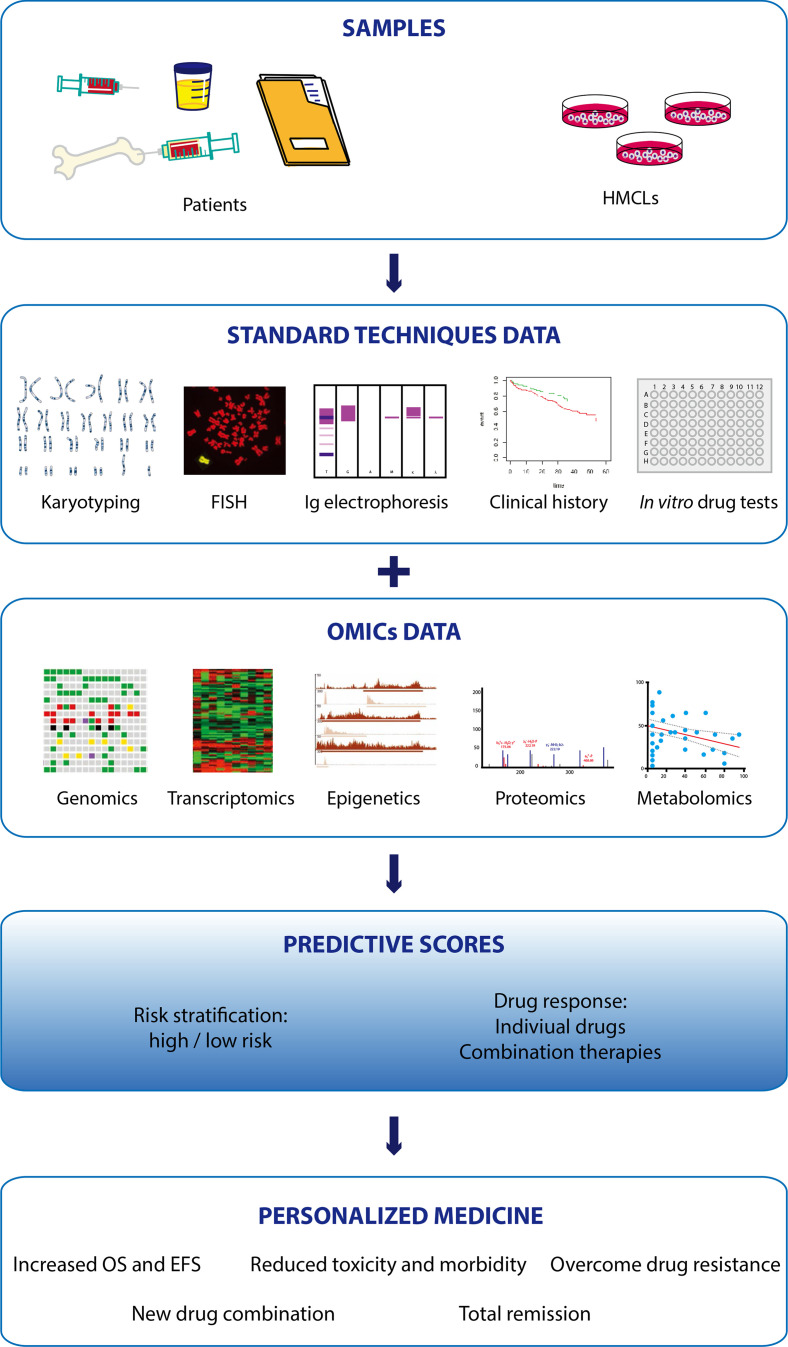
Figure 1.
Predictive scores in MM. Samples from patients (blood, urine, BM aspirates) and HMCLs are commonly studied by classical diagnosis techniques. Karyotyping and fluorescence in situ hybridization allow the detection of CNA and translocations, respectively; electrophoresis detects Ig chains in serum and urine from patients; clinical history collects data about lines of treatment, relapse, progression free survival, overall survival, MRD, and co-morbidities, that are crucial when analyzing cohorts of patients. Primary cells from patients and HMCLs can be used to assess the efficacy of in vitro drug treatments and drug combinations in order to find new therapeutic approaches. The integration of data from these routine techniques with omics obtained data, specially from genomics and transcriptomics studies, allows to build scores (Tables 3 and 4) capable to stratify the risk or predict drug response, that are instrumental for personalized treatment. Combination of several scores will refined diagnosis and improve the monitoring of the evolution of the disease. Finally, since drug resistance is the main reason of relapse, personalized medicine based on omics-developed scores will allow to choose the best drug for each patient, increasing the probability of survival while reducing treatment-associated toxicity, which also translates in better quality of life. It includes a karyotyping cartoon (Karyotyping) taken from an open source: https://smart.servier.com/smart_image/karyotype/