Figure 2. Identification of cannabinoid receptor 1 (CB1) inhibitor by high-throughput virtual screening and molecular docking.
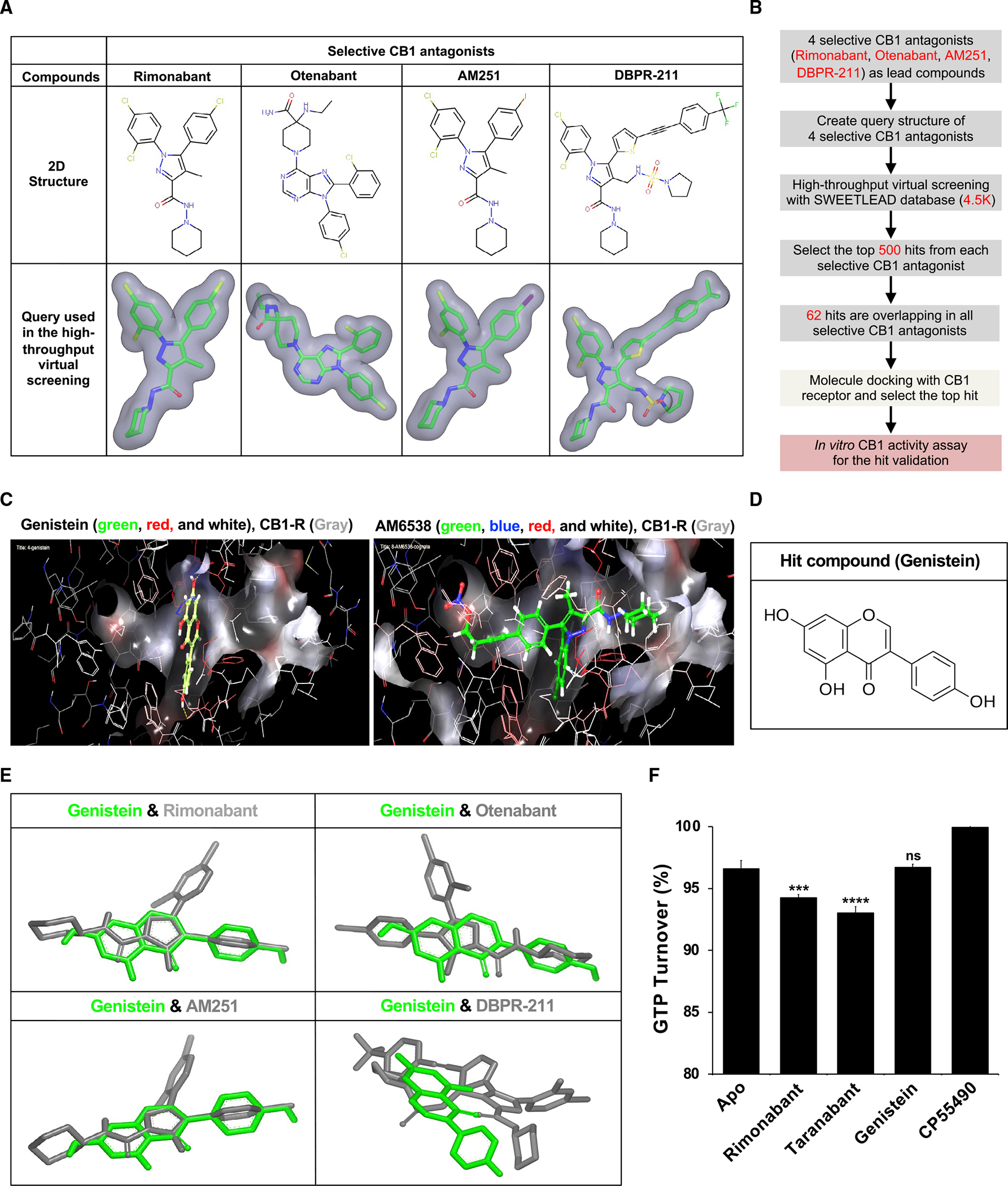
(A) Query structure of 4 selective CB1 antagonists (rimonabant, otenabant, AM251, and DBPR-211) was used in the ligand-based high-throughput virtual screening.
(B) High-throughput virtual screening workflow for lead compound identification.
(C) Docking of the selective CB1 antagonist AM6538 and genistein into the CB1 receptor. The best fit of AM6538 and genistein into CB1 was shown using Schrödinger molecule docking software.
(D) Molecular structure of hit compound genistein.
(E) Genistein shared structural homology with selective CB1 antagonists in ligand-based virtual screening.
(F) Genistein is a neutral antagonist of the CB1 receptor. GTPase-Glo assay reveals rimonabant and taranabant to decrease GTP turnover compared with Apo and GTPase without ligand, indicating that they are inverse agonists. Genistein elicits the same GTP turnover as Apo, suggesting that genistein is a neutral antagonist. CP55490 causes increased GTP turnover consistent with its function as a CB1 agonist. ***p < 0.001 versus Apo; ****p < 0.001 versus Apo; ns, not significant versus Apo.
