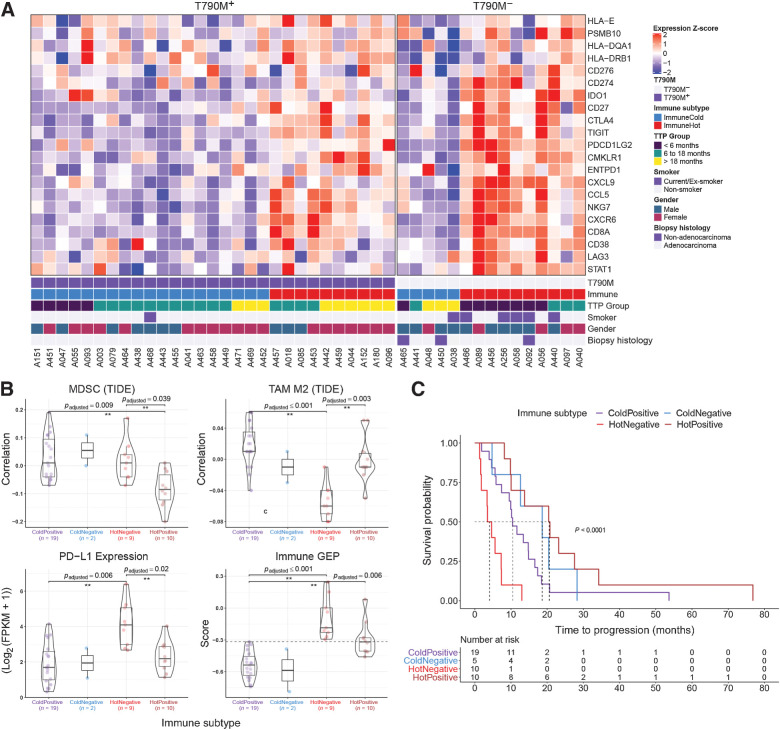
Figure 3.
Stromal transcriptomic correlates of EGFR TKI resistance. A, Relative expression of genes used in the immune GEP calculation (39). Patients were clustered into two groups using consensus k-mean clustering and sorted by T790M status followed by time to progression (TTP). B, Comparison of immune-suppressive cells correlation index (derived using TIDE) and expression levels of immune checkpoint genes (PD-L1 gene expression shown here) between immune subtypes. Immune GEP score was calculated using the method from Cristescu et al. (39). Horizontal line demarcates GEP score of −0.318, defined as the cutoff for high GEP in the original article. Pairwise comparison test was carried out using Games–Howell test and P values were adjusted using Benjamin–Hochberg procedure. C, Kaplan–Meier curve of EGFR TKI TTP comparing different immune subtypes.