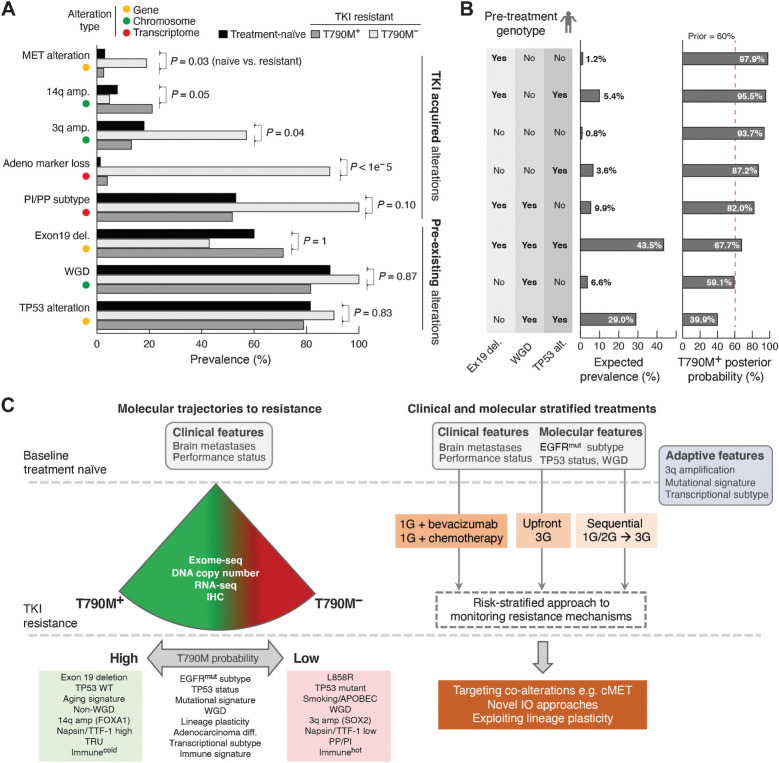
Figure 4.
Data-driven TKI treatment algorithm. A, Genomic and transcriptomic alterations with distinct frequencies in patients with T790M+ and T790M− disease. The observed prevalence of each alteration in T790M+ and T790M− groups as well as patients with treatment-naïve late-stage EGFR-mutated tumors are shown. Testing the null-hypothesis that frequencies of individual alteration types are not different between treatment-naïve and resistant cohorts, alterations were divided into either likely pre-existing or likely TKI-treatment acquired alterations. B, The expected patient prevalence for each (n = 8) combination/genotype of the three inferred pre-existing alterations. The posterior probability was estimated for each genotype using Bayesian updating. C, Summary of molecular features that modify probability of T790M (left), and potential to use baseline clinical and molecular features, as well as adaptive changes to determine optimal therapeutic strategy (right).