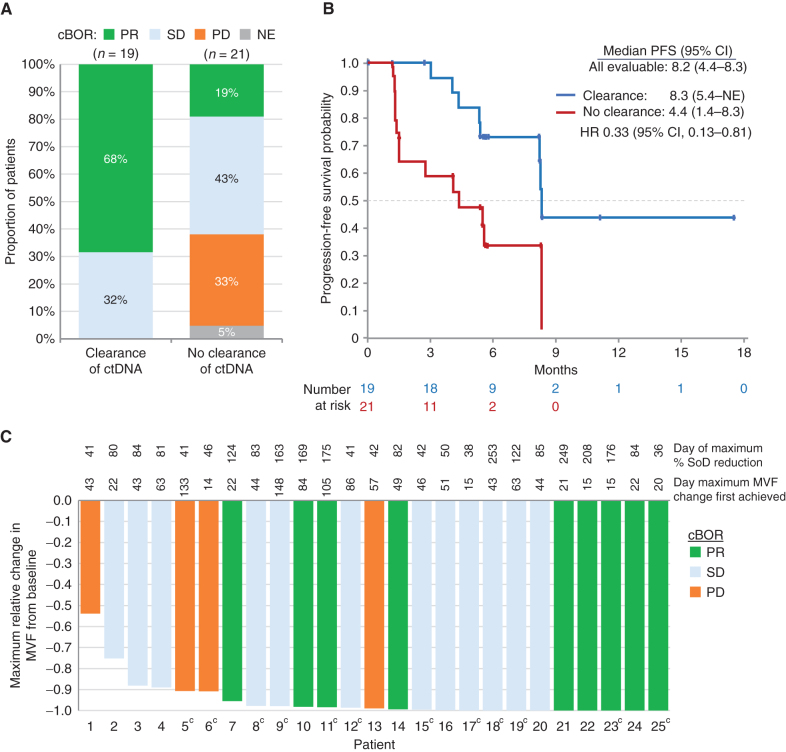
Figure 4.
Analyses of ctDNA changes. A, Proportions of confirmed BOR (cBOR; BICR per RECIST 1.1) by early clearance of ctDNA.aB, Kaplan–Meier plots showing PFS by early clearance of ctDNA.aC, Waterfall plot showing maximum change relative to baseline in the minor variant frequency (MVF) of EGFR-activating mutations in ctDNA and confirmed BOR by BICR (25 of 57 patients were evaluableb). aEarly clearance of ctDNA was defined as nondetectable plasma of both EGFR Ex19del and EGFR L858R at week 3 or week 6, where one or more allele comprising EGFR Ex19del or EGFR L858R was detectable at baseline. Serial ctDNA samples were collected from 45 of 57 patients, but 5 did not have evaluable ctDNA data for either week 3 or 6. In the 17 patients not evaluable for early clearance of ctDNA, responses were CR/PR, 29%; SD, 24%; PD, 17%; and NE, 35%. bFewer patients (25 of 57) were evaluable for relative change in ctDNA than for early clearance of ctDNA (40 of 57), as the former required ctDNA levels to be above the limit of quantification (MVF > 1%); the latter required levels to be above the limit of detection, which was lower (MVF > 0.02%). cPatients without GeneStrat data at C1D1 were analyzed if they had GuardantOMNI data at C1D1 when the MVF was >1%; this is because GeneStrat and GuardantOMNI data were concordant, except for the low MVF range (<1%). SoD, sum of diameters.