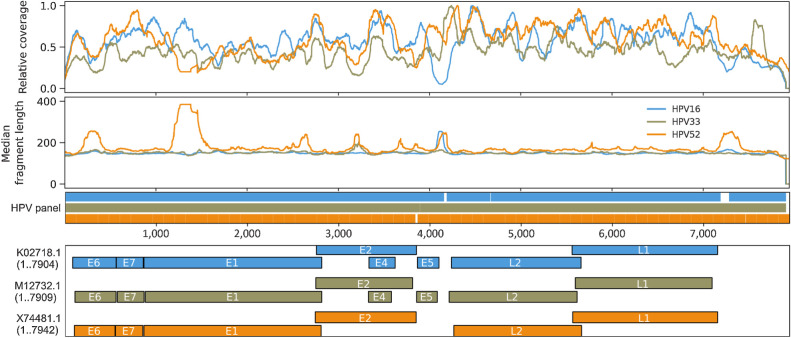
Figure 6.
Genome-wide fragmentomic patterns of HPV-mapping ctDNA fragments across 3 HPV genotypes. First track: Relative read depth coverage for HPV-mapping paired-end reads. Coverage is normalized relative to the maximum for each genome. Relative coverage of 0 indicates no properly paired mapping reads. Samples from tumors harboring HPV-16 (blue; N = 22 samples from 21 patients), HPV-33 (gold; N = 2 samples from 1 patient), and HPV-52 (orange; N = 2 samples from 1 patient) are shown. Second track: Median HPV ctDNA fragment lengths (bp) inferred from sequenced fragment insert sizes. Third track: Capture probes for the 3 HPV genotypes. Fourth track: HPV gene organization from GenBank.