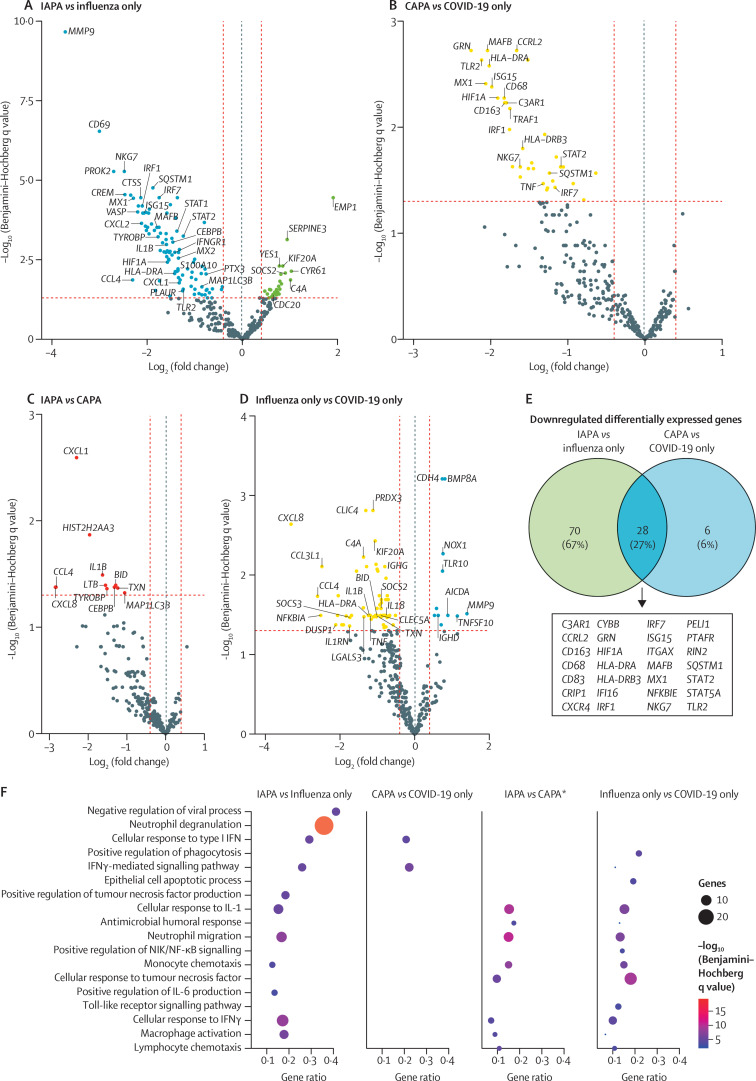
Figure 2.
Differentially expressed genes in bronchoalveolar lavage fluid samples from patients with IAPA, influenza only, CAPA, or COVID-19 only
Volcano plots showing differentially expressed genes for the comparisons of IAPA versus influenza only (A), CAPA versus COVID-19 only (B), IAPA versus CAPA (C), and influenza only versus COVID-19 only (D). A dotted blue line marks a log2(fold change) value of zero. A dotted red line crossing the y-axis marks a Benjamini–Hochberg q value of 0·05. The two dotted red lines crossing the x-axis marks a log2(fold change) of 0·4 and –0·4, respectively. We selected genes to present if they were linked to interesting pathways related to antiviral and antifungal immunity. (E) Venn diagram showing the downregulated differentially expressed genes shared in the comparisons of IAPA versus influenza only and CAPA versus COVID-19 only. (F) Dot plot showing ClueGO hypergeometric enrichment pathway analysis based on the downregulated differentially expressed genes of the four disease comparisons, with Gene Ontology biological process as the pathway database. *For the comparison of IAPA versus CAPA, differentially expressed genes with q values (Benjamini–Hochberg) of less than 0·20 were included to generate the analyses. Differentially expressed genes with q values (Benjamini–Hochberg) of less than 0·05 were used for all other comparisons. CAPA=COVID-19-associated pulmonary aspergillosis. IAPA=influenza-associated pulmonary aspergillosis. NIK=NF-κB-inducing kinase.