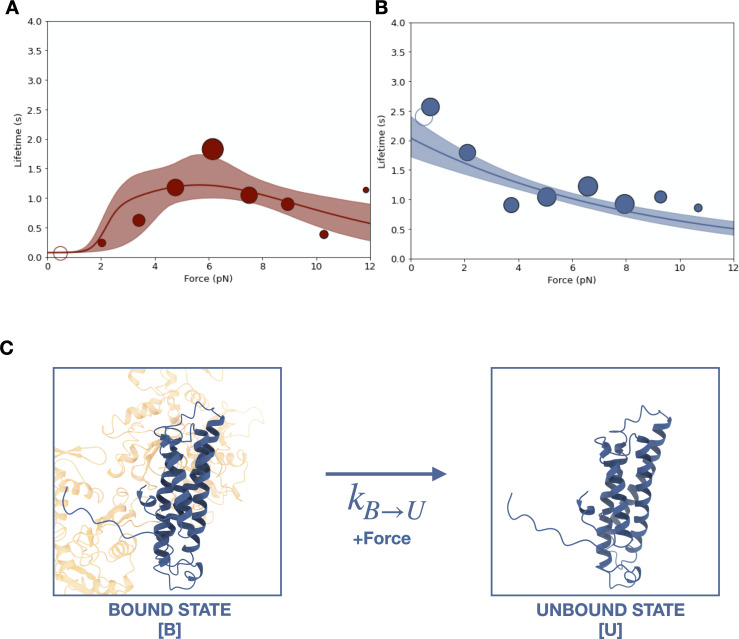
Figure 5. Force-dependent binding of cadherin/catenin complexes to F-actin.
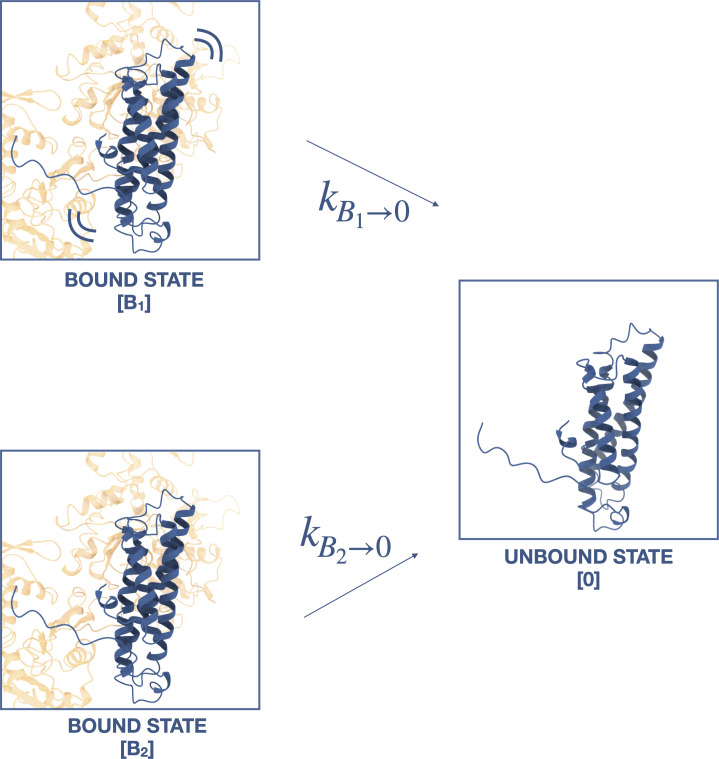
(A) Mean binding lifetimes (red filled circles) from constant-force assay measurements from previously reported (Bax et al., 2022) wild-type E-cadherin/β-catenin/αE-catenin complex data (N = 700). These data are represented here without depicting the direction of force applied relative to the polar actin filament, and fit to a nondirectional two-state catch bond (red curve). Unfilled circles represent the mean lifetime of events collected in the low-force assay (N = 90). Envelopes indicate 95% confidence intervals for the fit, obtained by empirical bootstrapping. Areas of all circles are proportional to the number of events measured in each equal-width bin. (B) Mean binding lifetimes (blue filled circles) from pooled low- (N = 145) and constant-force assay (N = 856) measurements for ternaryΔH1 complexes. These data were fit to a one-state slip bond model (blue curve). (C) The one-state slip bond model. The conformation of a bound actin-binding domain (ABD) missing H1, denoted state B, is comparable to the strong state of the two-state catch bond model. Molecules transition between bound (B) and unbound (U) states, where the dissociation rate, , increases exponentially with force.