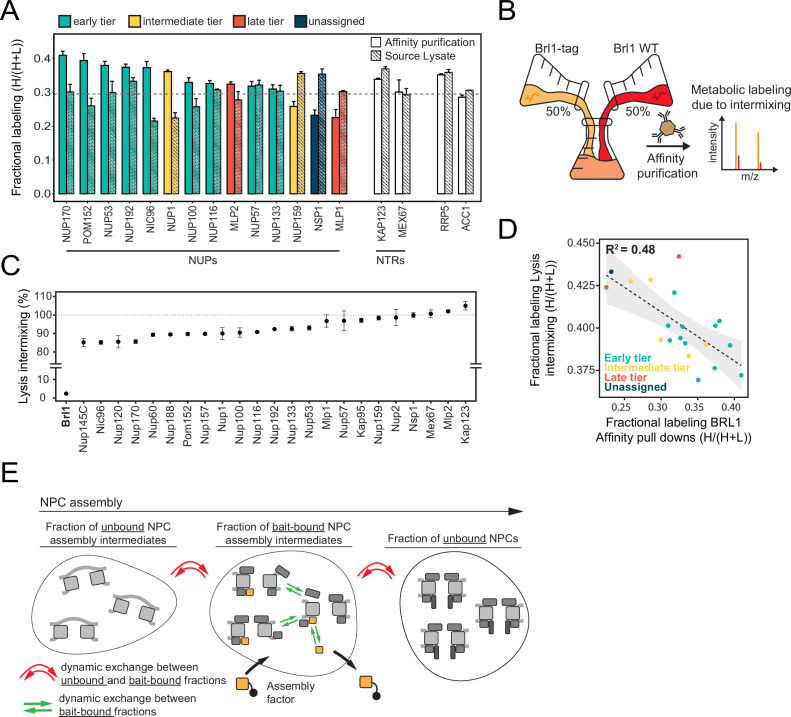
Appendix 1—figure 1. Control of protein labeling in KARMA (Kinetic Analysis of Incorporation Rates in Macromolecular Assemblies) assays.
(A) Fractional labeling of nucleoporins (NUPs) and nuclear transport receptors (NTRs) in KARMA assays with Brl1 bait and the respective source lysate, 90 min after labeling onset. Median ± SD of three biological replicates. (B) Experiment to test the intermixing dynamics. Equal fractions of an unlabeled Brl1 affinity-tagged strain and a wildtype culture grown in metabolically labeled medium were subjected to the affinity purification procedure. (C) Percentage of intermixing for NUPs and NTRs normalized to the mean of all co-purified proteins with Brl1 bait. Median ± SD of three biological replicates. (D) Correlation of NUPs between fractional labeling in the intermixing experiment and in KARMA assay with Brl1 bait. Coloring according to the assembly tier. Median of three biological replicates each. (E) During complex assembly, proteins can dynamically exchange within the bait-bound fraction (green arrows) or between the bait-bound and unbound fractions (red arrows). The metabolic labeling in KARMA assays is only sensitive to dynamic exchange between bait-bound fractions and unbound fractions, whereas in the lysate intermixing assays both forms of exchange contribute to the observed labeling.