Figure 4.
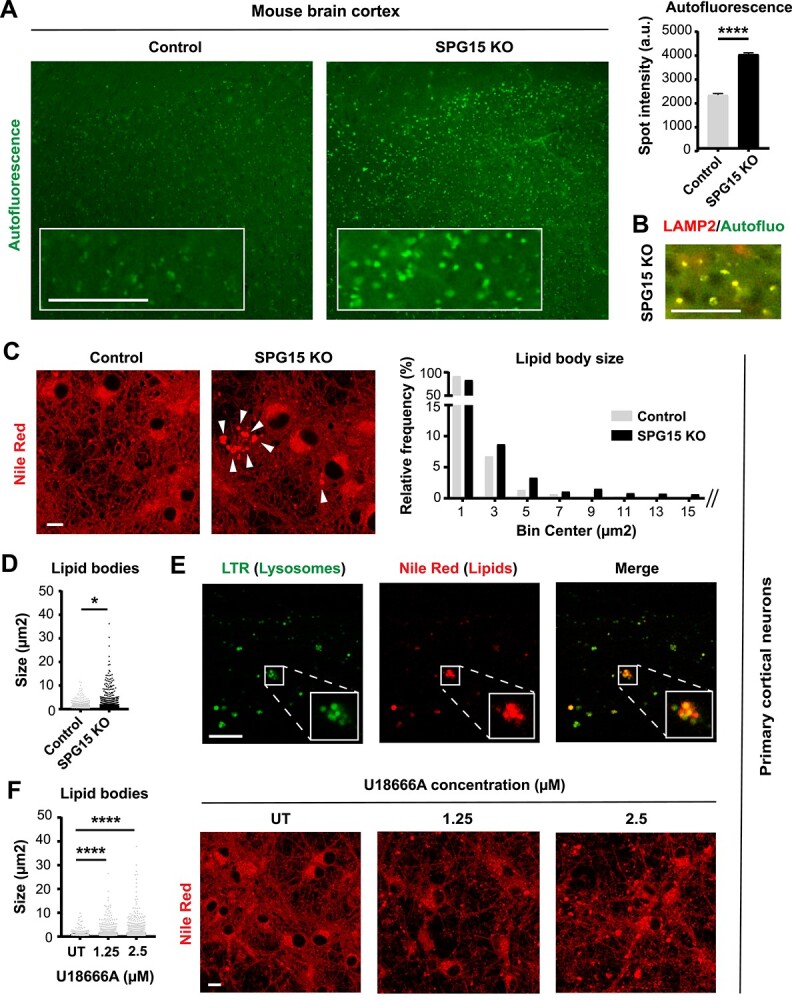
Loss of SPG15 impairs lysosomal lipid metabolism. (A) 15 month old SPG15 KO mice accumulate prominent autofluorescent particles throughout their cortex. Scale bar = 100 pixels. N = 3 (brains, with >1000 granules analyzed per brain). Mann–Whitney test, error bars represent SEM. **** corresponds to P < 0.0001. (B) Autofluorescent particles co-localize with LAMP2, suggesting that these structures are lipofuscin granules. Co-localization is evident from the yellow color generated by the overlap of red (LAMP2) and green (autofluorescence) pixels. Scale bar = 100 pixels. (C) Loss of SPG15 protein promotes the accumulation of lipid bodies (white arrowheads) in cultures of embryonic primary cortical neurons. Lipids were stained with Nile Red. Scale bar = 10 μm. The histogram shows the frequency distribution of lipid bodies binned as per size (μm2). The size of each individual lipid droplet was also plotted separately to highlight a significant difference between these populations (D). N = 3 (independent experiments, each evaluating three fields). t-test, * corresponds to P < 0.05. (E) Lipid bodies (stained with Nile Red) co-localize with lysosomes (stained with Lysotracker Green DND-26, LTR) as evident from the yellow color generated by the overlap between red and green signals. Scale bar = 10 μm. Note that these confocal micrographs represent the axonal side of cultures grown in a microfluidic chamber. (F) Treatment of control neuronal cultures with two doses of U18666A (1.25 or 2.5 μm) recapitulates the lipid droplet phenotype observed in SPG15 KO neurons, suggesting that loss of SPG15 protein prevents the egress of lipids from lysosomes. N = 3 (independent experiments, each evaluating three fields of view). t-test, **** corresponds to P < 0.0001. Scale bar = 10 μm. UT = untreated.
