Figure 3.
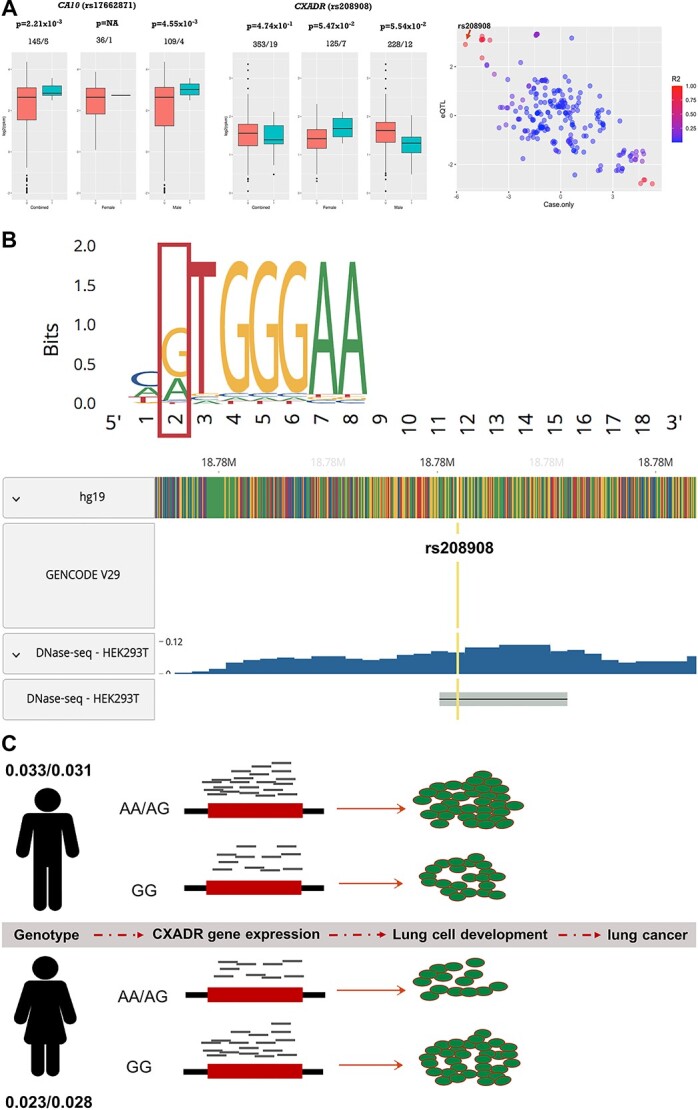
Functional analysis of identified variants. (a) eQTL analysis at candidate variants. Left and middle, box plot of gene expression from lung between individuals with none (0) and at least on risk allele (1) in female and male combined, female and male groups for CA10 and CXADR, respectively. The data from samples with Caucasian ancestry in GTEx were used in the analysis. Individuals with rpkm < 0.25 were removed from the analysis. P-values and number of samples were labeled above each plot. rs208908 was genotyped in GTEx; rs17662871 was imputed in SNP and SNP dosage ≤0.4 was coded as 0; SNP dosage ≥ 0.6 was coded as 1. Linear regression analysis of gene–sex interaction in predicting CXADR gene expression displayed P = 3.85×10−3. Right, scatter plot of z-scores from 229 SNPs, ranging from 18 745 702 to 18 806 105 (hg19) on chromosome 21 was displayed on the plot. X-axis represented gene–sex interaction analysis in lung cancer risk; and y-axis represented the z-scores from gene–sex interaction analysis in CXADR gene expression prediction. LD r2 was computed for each of the SNPs with rs208908 as reference. (b) Functional inference of rs208908. The predicted binding motif at rs208908 (highlighted in red color) for TF using PROMO (Upper). DNase-seq analysis in HEK293T (Human embryonic kidney) cells detected a peak covering rs208908 suggesting it was located within a cis-regulatory DNA sequence element (RegulomeDB bottom). (c) Presumptive disease risk model at rs208908. This variant, located upstream of CXADR, may affect lung cancer risk through gene expression regulation. The numbers indicate MAF for rs208908 in cases/controls between male and female population.
