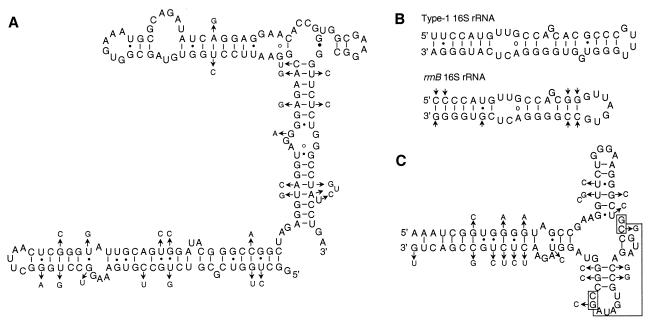
FIG. 3.
Compensating covariation of interacting bases. The secondary structures were generated according to the consensus models for 16S and 23S rRNAs (12). (A) Region corresponding to nt 588 to 753 of E. coli 16S rRNA (2). The nucleotide sequence of T. chromogena rrnF is presented to form the skeleton, and the smaller letters on the side denote the varied nucleotides in the rrnB sequence. Short bars connect the canonical pairings, such as G-C and A-U; small solid dots denote the noncanonical G-U and U-G pairings. Conserved A-G pairings are indicated by circles, and a G-G pairing is indicated by a large solid dot. The arrows point to covaried bases or base substitutions that lead to a different type of pairing, such as G-C to G-U. (B) Region of the 16S rRNA of T. chromogena rrnF and rrnB corresponding to nt 1118 to 1155 of the E. coli gene. This region of rrnB contains an eight-base deletion in comparison with rrnF. (C) Region of 23S rRNA of rrnB (nt 281 to 359, E. coli numbering) that differ from the corresponding region of rrnF at 32.5% nucleotide positions. Two pairs of bases involved in tertiary interaction are boxed and linked by a line.