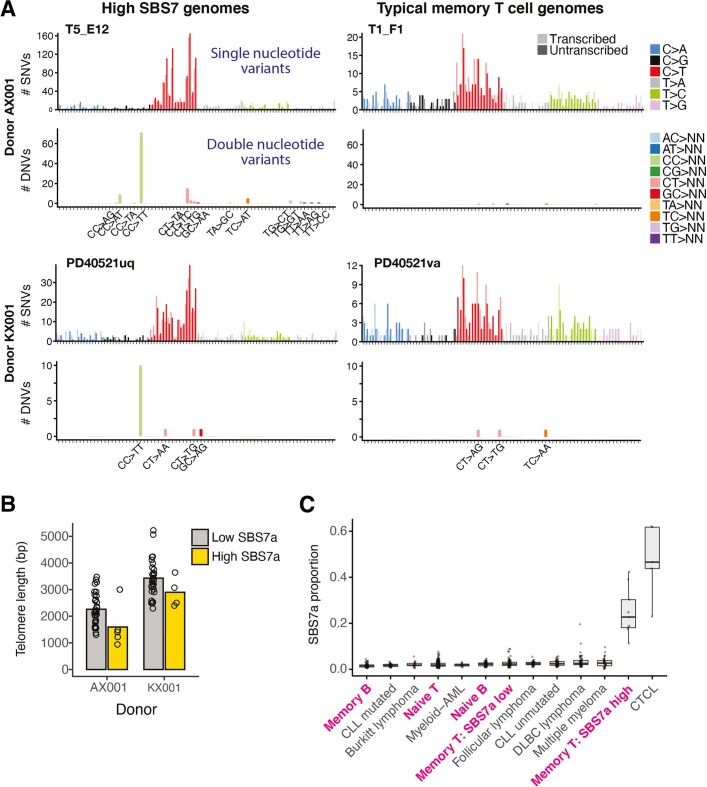
Extended Data Fig. 5. Ultraviolet light mutational signature (SBS7a) in lymphocytes.
(A) Raw mutational spectra shown for all mutation calls from four lymphocyte colonies, two with high contribution of SBS7a (left) and two with a more typical T-cell spectrum (right) from two different donors (rows). For each cell, the top panel shows the SNV spectrum, with trinucleotide contexts on the x-axis representing 16 bars within each substitution class, divided into 4 sets of 4 bars, grouped by the nucleotide 5′ to the mutated base, and within each group by the 3′ nucleotide. The bottom panel shows frequency of dinucleotide substitutions. (B) Telomere lengths for memory T cells with (yellow) and without (grey) high SBS7a signature. A memory T cell with high UV signature is defined as having greater than 9.5% (2 standard deviations above the mean) of its mutations attributable to SBS7a. (C) Proportion of mutations attributable to SBS7a across normal lymphocytes (paediatric samples excluded) and lymphoid malignancies. Boxes show the interquartile range and the centre horizontal lines show the median. Whiskers extend to the minimum of either the range or 1.5× the interquartile range. Number of genomes included per group: naive B: 68, memory B: 68, naive T: 332, memory T SBS7a low: 78, memory T SBS7a high: 9, Burkitt lymphoma: 17, CLL (chronic lymphocytic leukaemia) mutated: 38, CLL unmutated: 45, C. (cutaneous) T-cell lymphoma: 5, DLBC (Diffuse Large B-cell) lymphoma: 47, follicular lymphoma: 36, multiple myeloma: 30, myeloid-AML (acute myeloid leukaemia): 10.