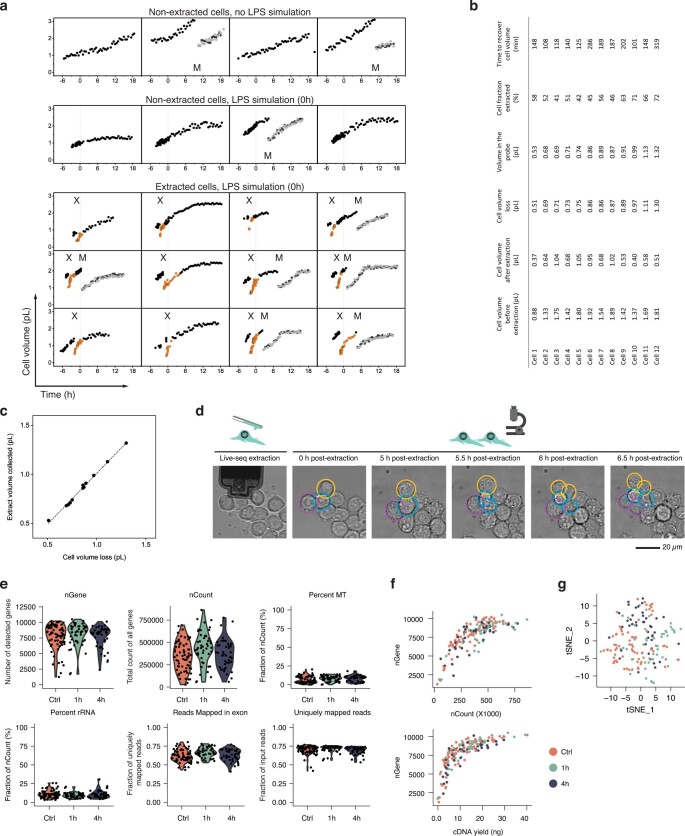
Extended Data Fig. 7. Live-seq preserves a cell’s growth, with additional controls relative to Fig. 3.
(a) Longitudinal measurements of RAW cell volumes. Cells were exposed to LPS at time 0 (vertical line). “X” indicates an extraction and “M” mitosis. After mitosis, both daughter cells were monitored (light and dark grey points). Volumes measured during cell recovery from Live-seq extraction are labeled as orange points. (b) The volume changes, extracted volumes, and recovery times of RAW cells, calculated based on the profiles in (a). (c) Correlation between the measured cell volume loss shown in (a) and the measured cell extract volumes that were measured in the FluidFM probe (see “Determination of the extracted volumes” section in Methods) (R2 = 0.99). (d) Representative time-lapse images showing the division of RAW cells post-Live-seq extraction. Three dividing cells are outlined in colored circles. The cell in the yellow circle was subjected to cytoplasmic extraction, while those in purple and blue were not. 49 extracted cells and 272 non-extracted cells were observed independently; 17 extracted and 54 non-extracted cells divided during 8 h of time-lapse imaging. (e) Quality control of the scRNA-seq data of the control IBA cells, as well as IBA cells 1 h and 4 h post Live-seq extraction, respectively. Relative to Fig. 3. (f) Plotted correlations between the number of detected genes on the one hand and respectively the total count of all genes (nCount, upper panel) and the cDNA yield (lower panel), were indistinguishable between the respective cell categories. (g) A tSNE projection of control IBA cell scRNA-seq (Smart-seq2) data (Ctrl, 70 cells) as well as 1 h (49 cells) and 4 h (43 cells) post Live-seq extraction scRNA-seq (Smart-seq2) data does not reveal clearly distinct clusters based on the top 500 most variable genes.