Fig. 2 |. Schematic representation of the diverse AAA+ proteins that coordinate eukaryotic protein quality control.
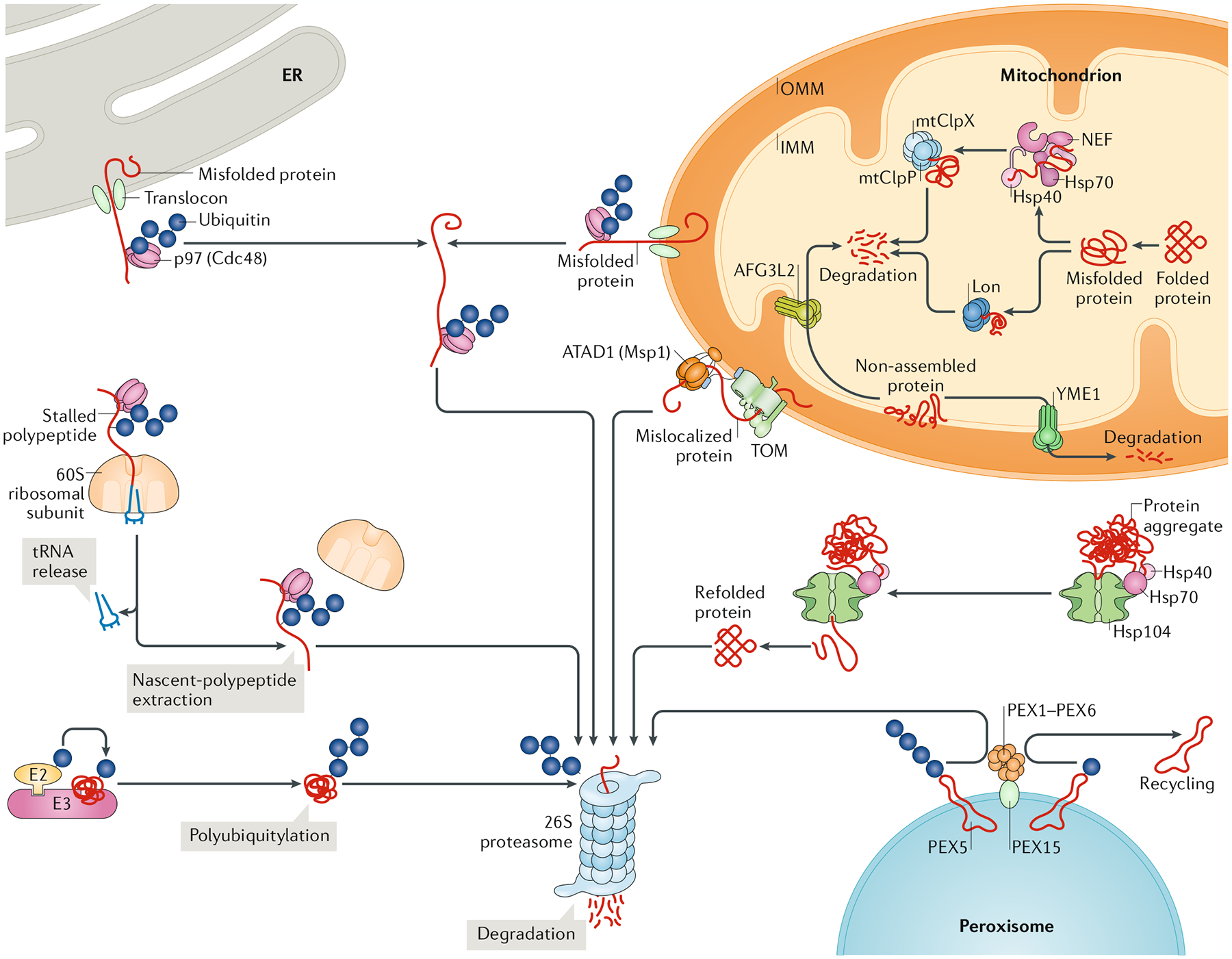
The 26S proteasome is the primary cellular machinery that unfolds and degrades ubiquitylated substrates and constitutes a central component of the protein quality control system, the main function of which is the removal of aberrant (for example, misfolded, truncated or damaged) proteins to maintain protein homeostasis (proteostasis). The 26S proteasome is a complex of numerous adaptor and regulatory components that assemble around a heterohexameric ATPase associated with diverse cellular activities (AAA+) motor, which inserts into a proteolytic barrel. The ubiquitin–proteasome system relies on the activity of E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases for tightly regulated ubiquitylation of substrates targeted for proteasome-mediated degradation. This pathway operates in the cytoplasm and in eukaryotes also in the nucleus (not shown). Multiple other AAA+ proteins cooperate with the 26S proteasome in protein quality control. Type II AAA+ proteins p97 (Cdc48 in yeast) and hetero-oligomeric PEX1–PEX6 extract polyubiquitylated substrates from the endoplasmic reticulum (ER) and peroxisomal membranes, respectively, for subsequent proteasomal degradation in the cytosol. In the mitochondrial outer membrane (OMM), the type I AAA+ protein ATAD1 (Msp1 in yeast) powers retrotranslocation of mistargeted proteins into the cytosol for degradation. Misfolded OMM proteins are retrotranslocated by p97 in a manner equivalent to ER-associated degradation. p97 is also involved in degrading aberrant polypeptides from stalled ribosomes during ribosome-associated protein quality control. In addition to the activity of the proteasome–ubiquitin system, independent proteolytic systems operate inside mitochondria. Here, AAA+ proteases YME1 and AFG3L2 from the classical clade, as well as ClpXP and Lon from the HCLR clade, degrade aberrant proteins in the inner mitochondrial membrane (IMM) and matrix, respectively. In addition to protein degradation, protein quality control relies on the capacity to refold misfolded proteins. This is mediated by chaperone complexes, including type II AAA+ disaggregases heat shock protein 104 (Hsp104; in yeast) and ClpB (in bacteria, mitochondria and plastids; not shown) that solubilize misfolded proteins and their aggregates, allowing their refolding. mt, mitochondrial; NEF, nucleotide exchange factor; TOM, translocase of the outer membrane.
