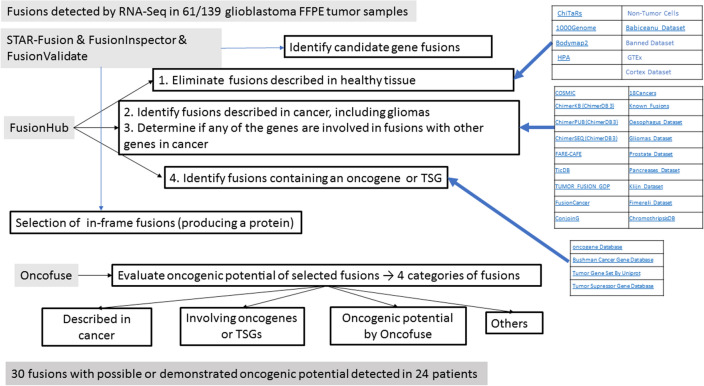
Figure 2.
Procedures and data bases used in the present study to select the fusions with oncogenic potential. From the long list of fusions detected by RNA-Seq, we used STAR-Fusion to detect fusion genes and FusionInspector to validate predicted fusions. We then used FusionHub to eliminate fusions previously described in healthy tissue, identify fusions previously described in cancers, and explore whether either gene had been identified as an oncogene or tumor suppressor gene (TSG) or had been associated with cancer. We next used FusionValidate to select only in-frame fusions and finally ran Oncofuse to predict the oncogenic potential of each fusion.