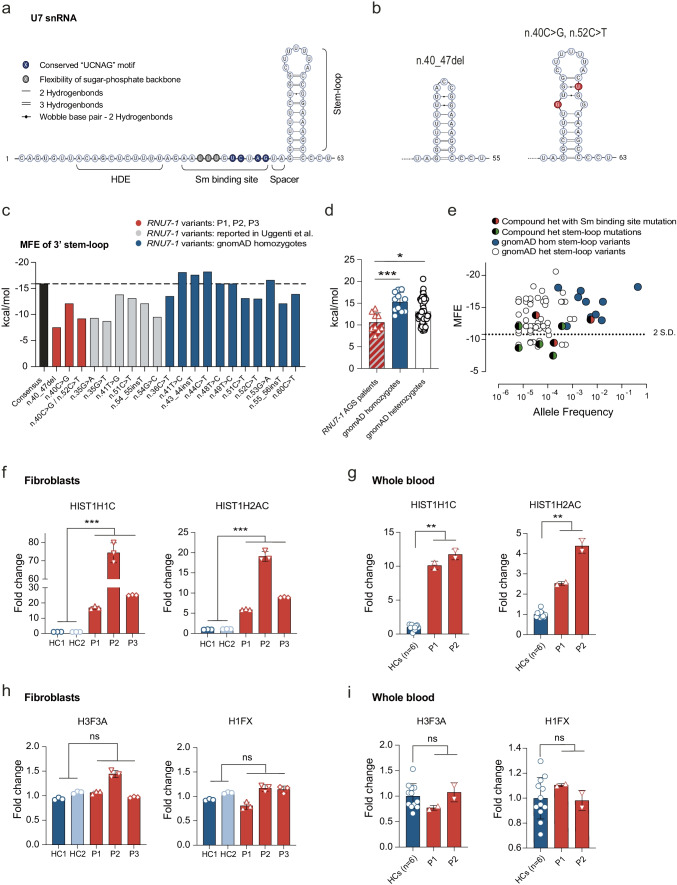
Fig. 2.
RNU7-1 mutations in the 3′ stem-loop weaken secondary structure and result in RDH pre-mRNA misprocessing. a Secondary structure and functional domains of U7 snRNA. Correct assembly of a functional U7 snRNP requires the flexibility of the sugar-phosphate backbone by uridines 23–25 (gray), a conserved “UCNAG” motif (blue) within the Sm binding site, spacing of two or more nucleotides between stem-loop and the Sm binding site, and a stable 3′ stem-loop. b Secondary structure of n.40_47del and n.40C > G/n.52C > T stem-loop mutations. c MFE of 3′ stem-loop consensus sequence (black), RNU7-1 variants of P1, P2, P3 (red), reported RNU7-1 mutations in AGS patients (gray), and homozygous carriers in gnomAD (blue). d MFE comparison of biallelic RNU7-1 mutations found in AGS patients compared to gnomAD homozygote and heterozygote RNU7-1 variants. Statistical analysis using the non-parametric unpaired Mann–Whitney U test (*** P < 0.001, * P < 0.5). Mean with s.d. is shown. e Graph of MFE against allele frequency for RNU7-1 stem-loop mutations in AGS patients, heterozygous and homozygous stem-loop variants in gnomAD. f, g Oligo(dT)-primed reverse transcription of polyadenylated RNA including replication-dependent linker (HIST1H1C) and core (HIST1H2AC) histones in primary fibroblasts (f) or whole blood (g) of RNU7-1 patients and HCs. h, i Expression of polyadenylated replication-independent histones (H3F3A, H1FX) in primary fibroblasts (h) or whole blood (i) of RNU7-1 patients and HCs. Data (f–i) presented as fold change normalized to cellular SDHA relative to the values for HCs. Data shown are representative for two (f–i) independent experiments with mean and s.d. of n = 3 technical replicates (f, h) or n = 2 technical replicates (g, i). **P < 0.01, ***P < 0.001, (f–i) by non-parametric unpaired Mann–Whitney U test