Figure 1.
Genome editing of genes with strong promoters
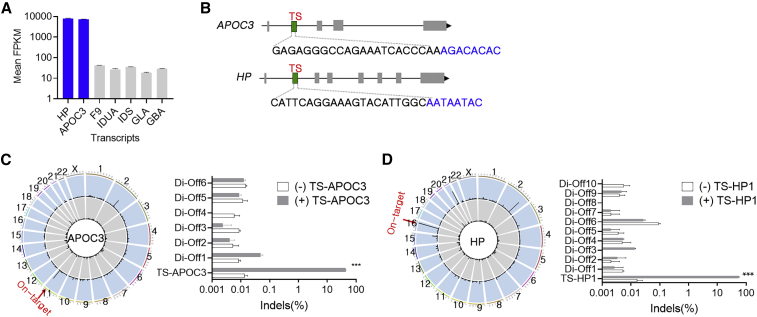
(A) RNA sequencing results from human primary hepatocytes (n = 3). (B) A schematic drawing of APOC3 and HP genes and the lead sgRNAs. Green regions in intron 1 of APOC3 and HP genes indicate target sites for the corresponding lead sgRNA, target site (TS)-APOC3 and TS-HP. TS-APOC3 and TS-HP are the same as TS7 and TS10, respectively in Figure S1. Blue text indicates the PAM. (C and D) Left, Genome-wide Circos plots showing in vitro cleavage sites in the human genome in the absence (gray) or presence (blue) of TS-APOC3 (C) or TS-HP sgRNA (D). On-target cleavage is indicated by the red arrow. Right, Targeted deep sequencing results for the off-target candidates obtained by Digenome-seq in TS-APOC3- (C) or TS-HP-treated HEK293 cells (D) (n = 3). Data are presented as means ± SEM, and statistical analysis was performed using the Student t test (A), (C), and (D). ∗∗∗p < 0.001.