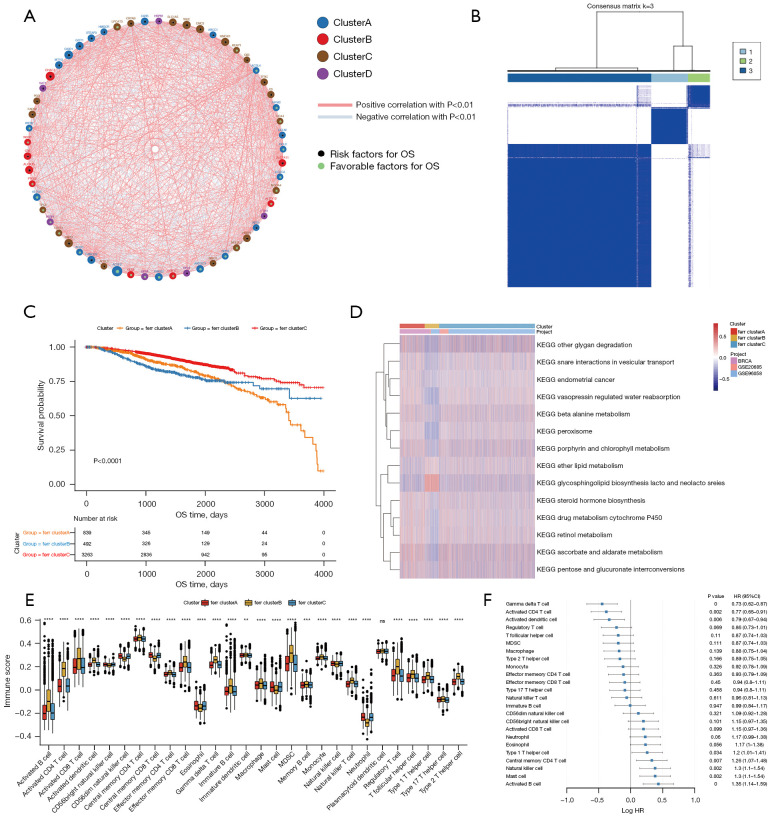
Figure 2.
Unsupervised clustering of ferroptosis genes in breast cancer samples. (A) The interaction between ferroptosis genes. The size of the circle indicates the effect of each gene on survival prediction, and the greater the expression, the more relevant the prognosis. Green dots in a circle represent prognostic protective factors, and black dots in a circle represent prognostic risk factors. The lines linking the genes show their interactions, and negative correlation is denoted by blue, while positive correlation is denoted by red. (B) Consistent clustering of ferroptosis genes, where 1, 2, and 3 represent the 3 ferr.cluster subgroups. (C) Kaplan-Meier curves show significant survival differences among the 3 ferr.cluster subgroups. (D) GSVA enrichment analysis demonstrating biological pathway activity in various ferr.cluster subgroups. Thermograms are used to visualize these biological processes, with red representing activation and blue representing inhibition. (E) The distribution and immune infiltration of 28 immune cell types in the 3 ferr.cluster subgroups. (F) An analysis of the prognosis of differential cells. In the figures, ns (not significant) stands for P>0.05, **, P<0.01; ***, P<0.001; ****, P<0.0001. GSVA, Gene Set Variation Analysis.