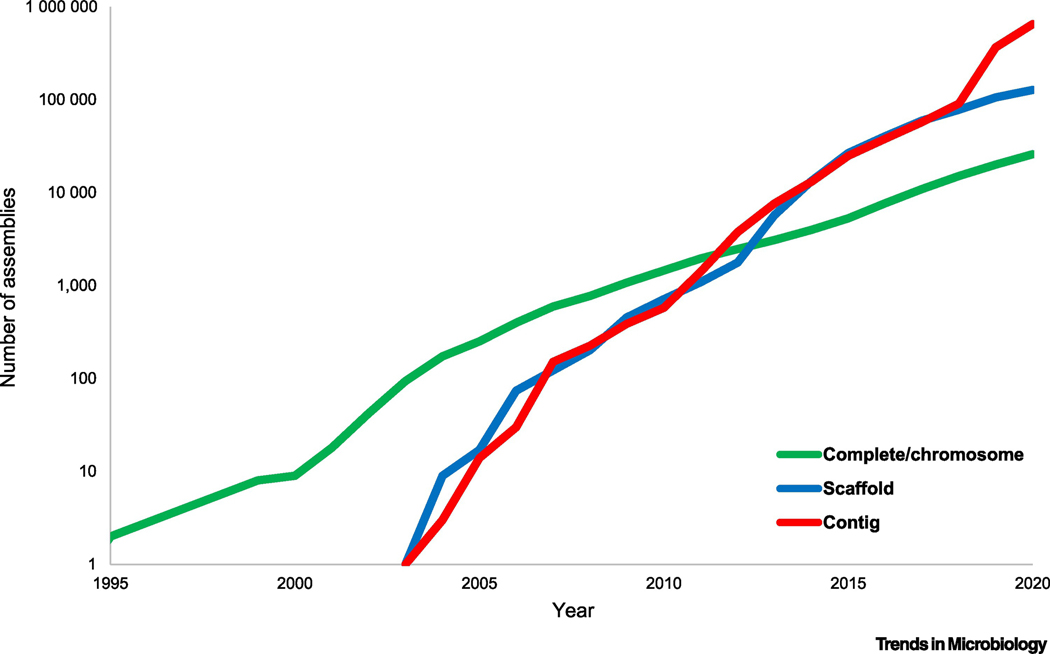
Figure 1. Exponential growth of the prokaryote genome database.
Number of genome assemblies at different assembly levels (ftp://ftp.ncbi.nih.gov/genomes/ASSEMBLY_REPORTS/) in the GenBank section of the NCBI Assembly database. The data points correspond to the end of the respective year; the X-axis starts with 1995, the year when the first two complete genomes were published.
Complete genome: all chromosomes are fully assembled with gaps not exceeding 10 ambiguous bases.
Chromosome: all chromosomes are fully assembled, but possibly containing gaps or unlocalized scaffolds.
Scaffold: sequence contigs are connected across gaps, but not placed on the chromosomes.
Contig: only unconnected contigs are available.