Figure 1.
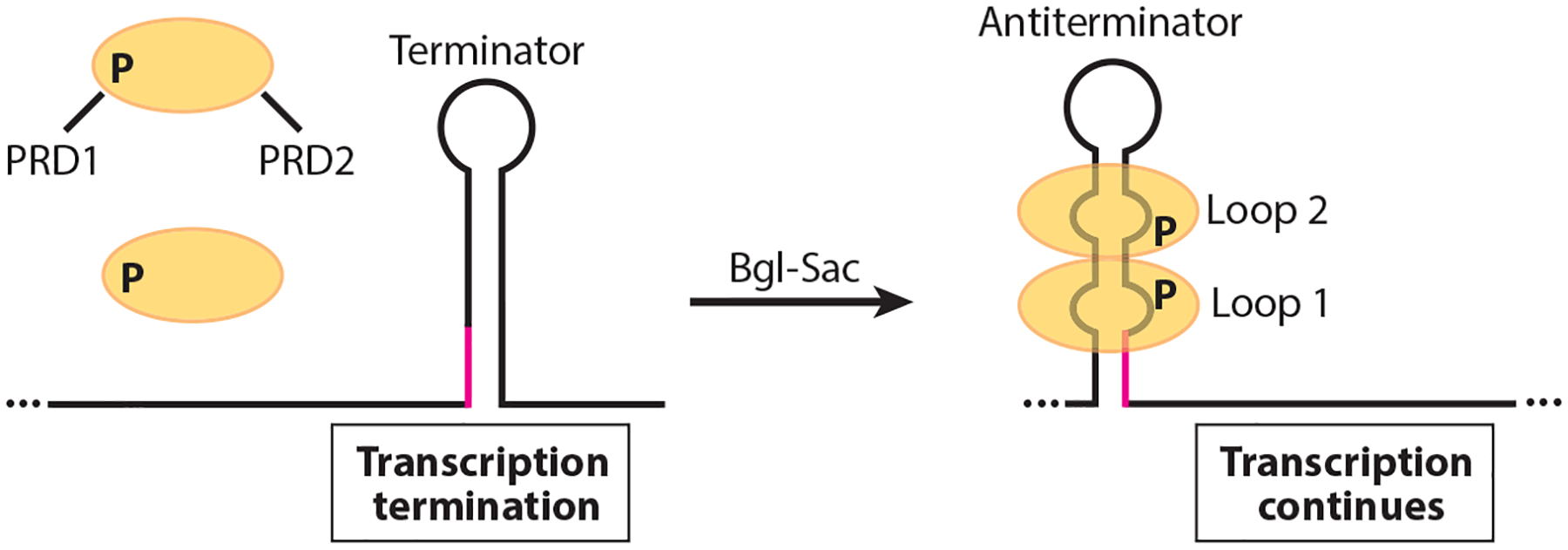
Model of the Bgl-Sac family antitermination mechanism. In the absence of the cognate carbon source the antitermination protein (yellow ovals) is rendered inactive by phosphorylation of PRD1. In the presence of the carbon source, PRD1 is dephosphorylated and PRD2 is phosphorylated, resulting in protein dimerization. The active dimer then binds to its cognate RNA target, thereby stabilizing an otherwise weak antiterminator structure. As a consequence, an overlapping intrinsic terminator is unable to form (overlap in magenta), leading to transcription of the operon involved in catabolism of the cognate carbon source. In the absence of bound protein, the terminator hairpin forms and transcription halts upstream of the coding sequence.
