Figure 3.
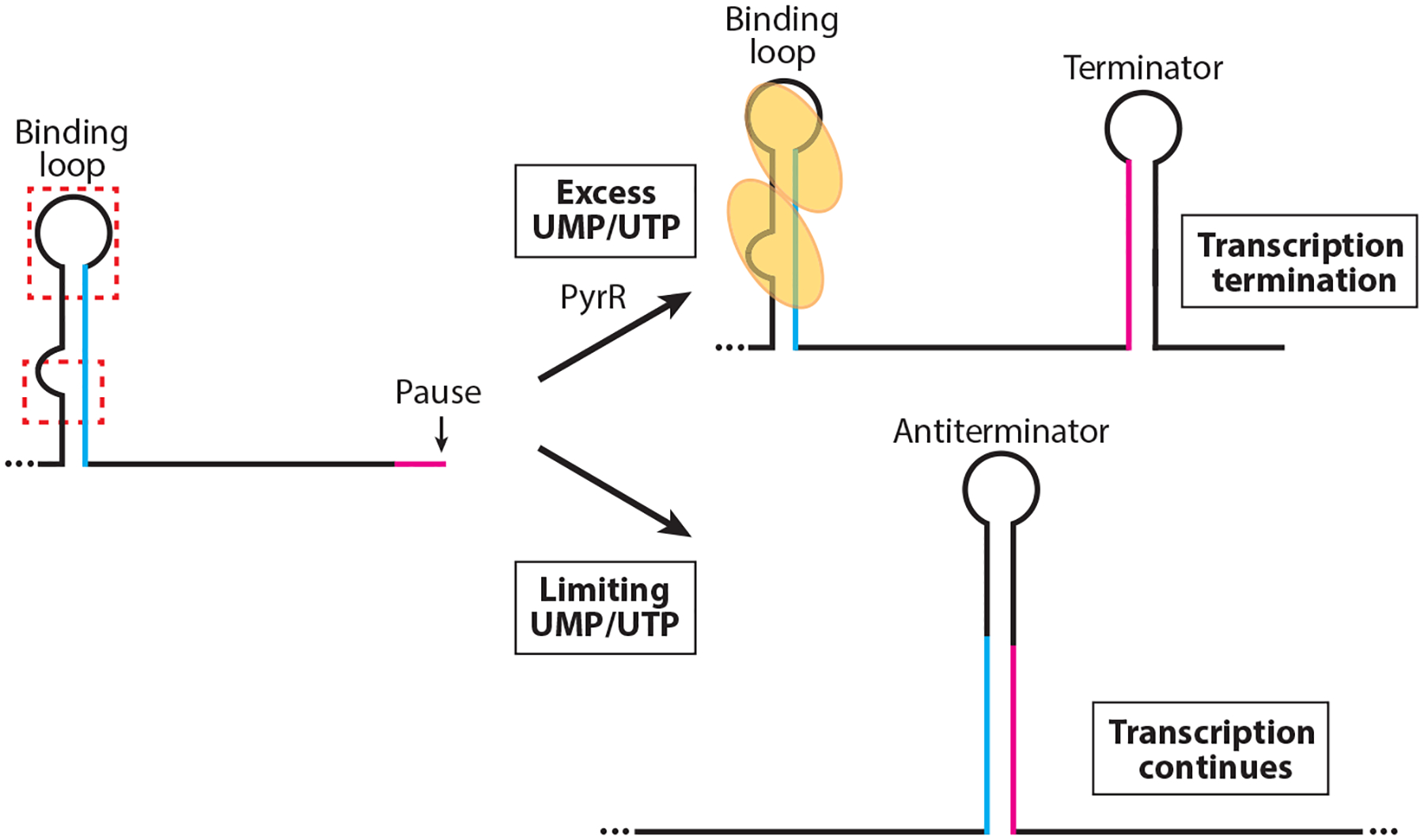
Model of the PyrR-mediated transcription attenuation mechanism. During transcription RNA polymerase pauses downstream of the PyrR-binding loop and within the overlap between the antiterminator and terminator structures (magenta). PyrR (yellow ovals) is not activated in limiting UMP/UTP conditions and does not bind to the binding loops in the 5′ UTR, or to the pyrR-pyrP and pyrP-pyrB intergenic regions. Once RNA polymerase resumes transcription, the antiterminator forms, which prevents formation of the mutually exclusive terminator hairpin (overlap in blue). In excess UMP/UTP conditions, UMP/UTP-activated PyrR binds to two regions of the binding loop (red dashed boxes). Pausing allows additional time for PyrR binding. Bound PyrR promotes transcription termination by preventing formation of the antiterminator structure.
