Figure 1.
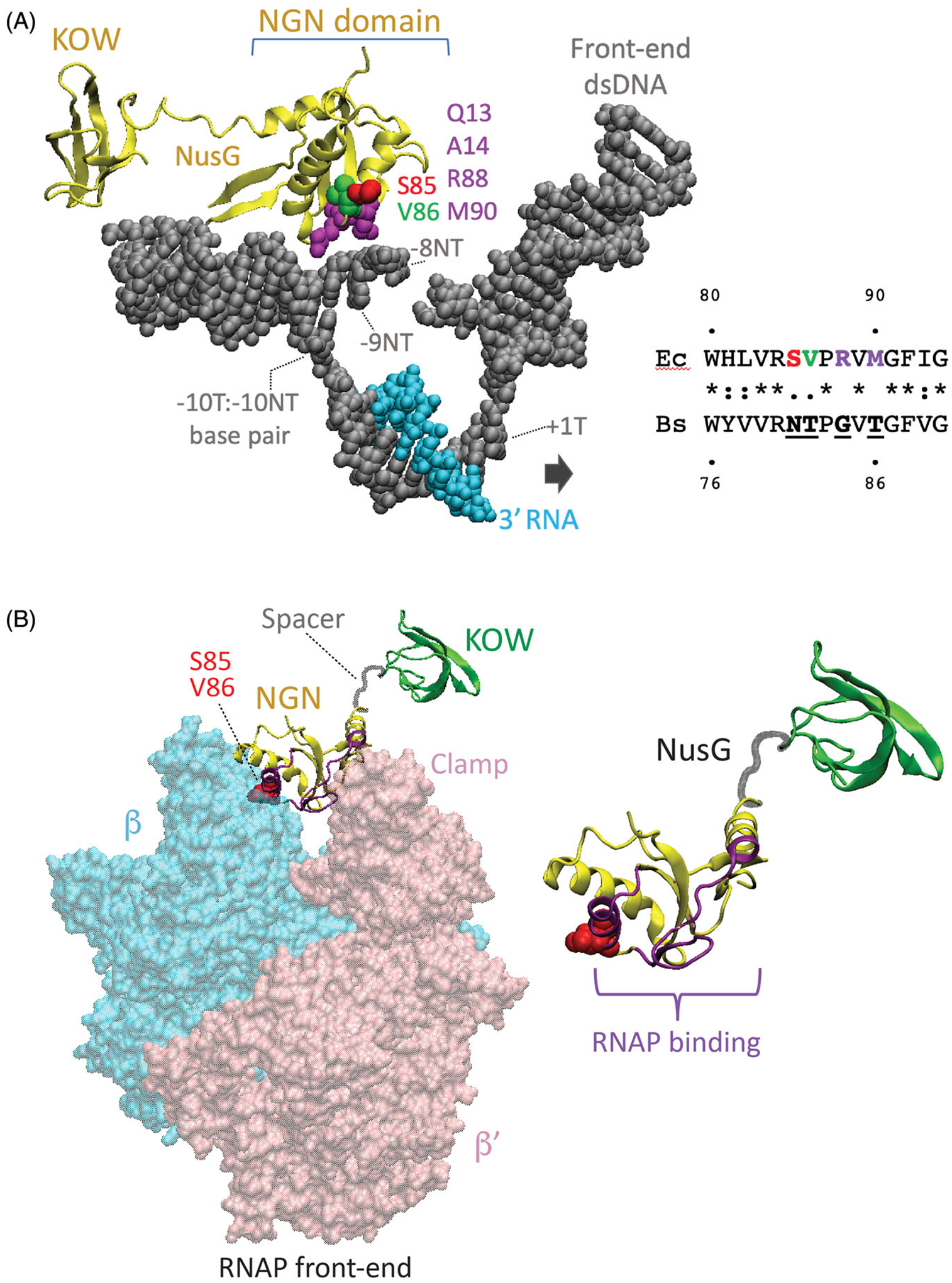
Structural foundation for NusG-dependent pausing. (A) Cryo-EM structure of the E. coli TEC containing Nus factors and N protein of bacteriophage lambda (PDB: 6GOV, Krupp et al. 2019). All Nus factors and RNAP subunits were removed for clarity. E. coli NusG shows the KOW and NGN domains with the NGN domain bound to a nucleic acid scaffold containing DNA (gray) and RNA (cyan). The residues in the template (T) and the non-template (NT) DNA strands in the vicinity of the NGN domain are shown with the numbers indicating their distance from the RNA 3’ end. Surface exposed residues of the NusG NGN domain in close proximity to the ntDNA strand within the transcription bubble are color-coded. Residues of the B. subtilis NGN domain that are vital for pausing (N81 and T82) correspond to S85 and V86 of E. coli NusG, respectively. (B) Binding site for the NGN domain on RNAP (PDB: 6GOV). NGN domain, yellow; KOW domain, green; Spacer, gray. Only β (cyan) and β’ (pink) subunits of RNAP are shown. Surface of the NGN domain and the S85/V86 residues at the interface with RNAP are colored in magenta and red, respectively.
