Figure 2.
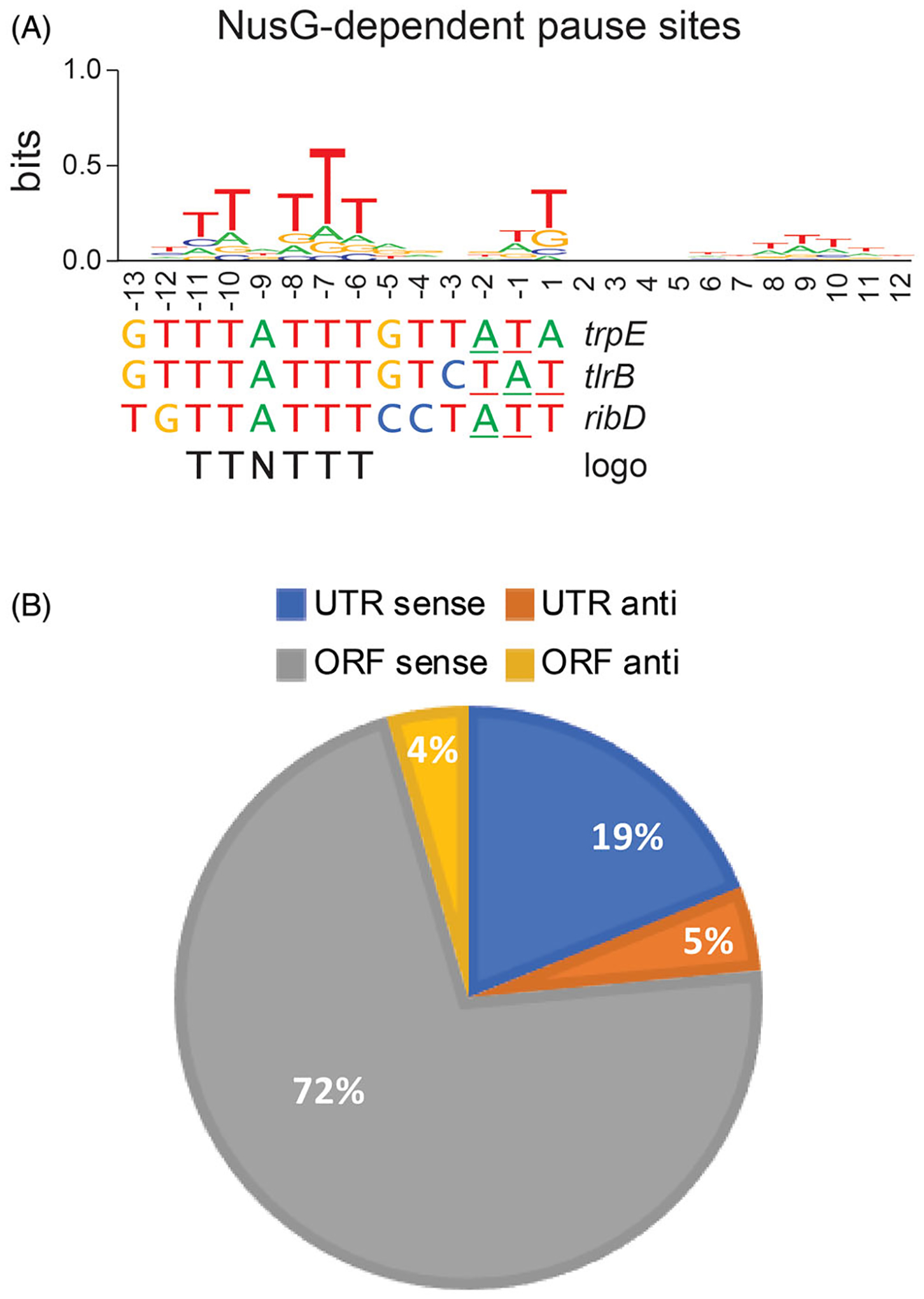
NusG-dependent pausing is sequence-specific and occurs throughout the B. subtilis genome. (A) TTNTTT sequence logo of the 1600 NusG-dependent pause sites identified in vivo by RNET-seq. This logo is shared by the trpE, tlrB and ribD pause sites that have been characterized in vitro. 3’ ends of paused RNA are underlined. (B) Genome-wide distribution of NusG-dependent pause sites in untranslated (UTR) and coding (ORF) regions in the sense (sense) and antisense (anti) directions.
