Figure 5.
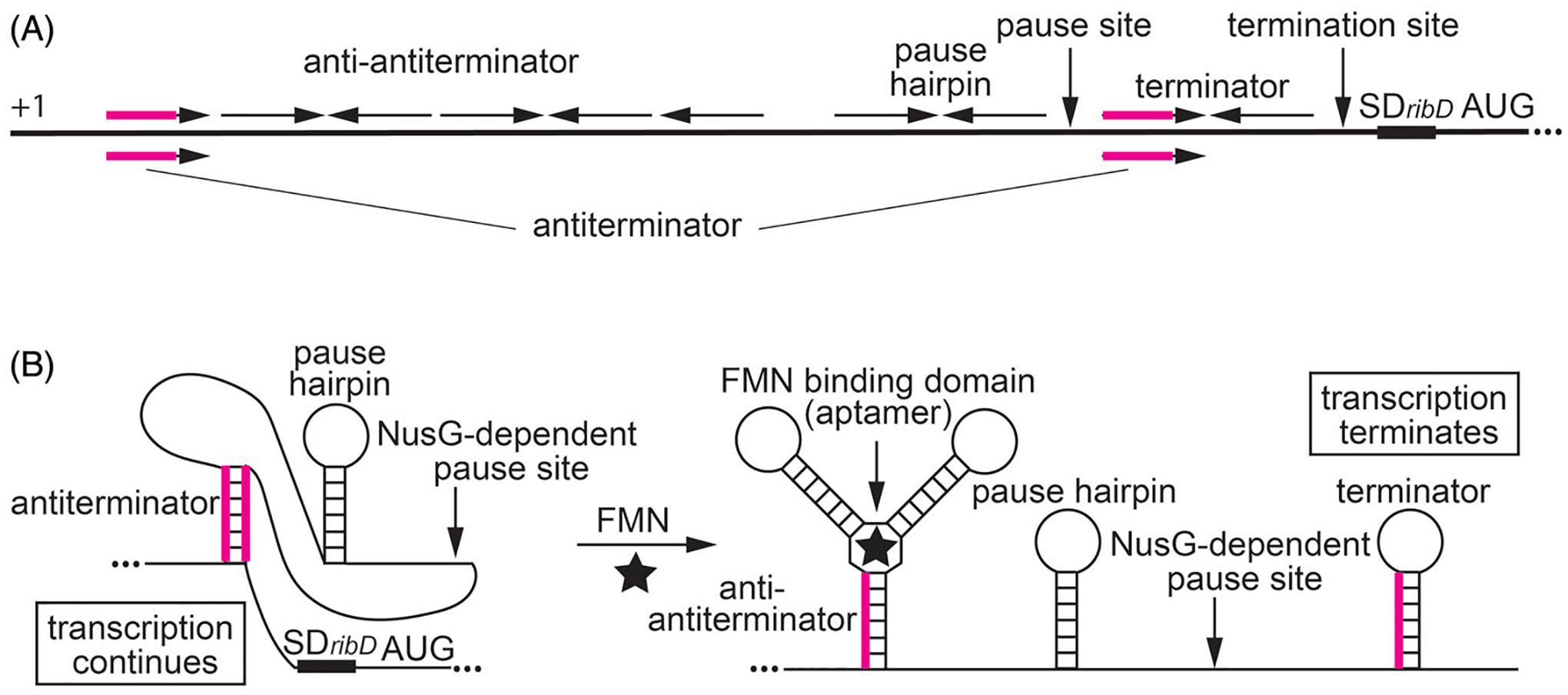
NusG-dependent pausing participates in the regulation of the ribDEAHT operon by an FMN-sensing riboswitch. (A) Schematic representation of the ribDEAHT leader region. The thick black line represents the leader region between the start of transcription (+1) and the AUG start codon for ribD. Inverted repeats for the anti-antiterminator, pause hairpin, and the terminator hairpin are shown above the line. The antiterminator that overlaps the anti-antiterminator and terminator structures is shown below the line (overlap in magenta). Positions of the NusG-dependent pause site, termination site, and the ribD SD sequence and AUG start codon are also labeled. (B) Model of FMN-dependent transcription attenuation. During transcription, RNAP pauses at a NusG-dependent pause site. Under FMN-limiting conditions (left), FMN does not bind to the nascent transcript. RNAP eventually overcomes the pause and resumes transcription. In this case, formation of the antiterminator structure prevents the formation of the terminator hairpin, resulting in transcription readthrough. Under FMN-excess conditions (right), FMN binds to the aptamer and stabilizes the anti-antiterminator structure, which prevents the formation of the antiterminator structure. As a consequence, formation of the terminator hairpin causes transcription to terminate. NusG-dependent pausing provides additional time for FMN binding and reduces the concentration of FMN required for termination. Color coding is the same as in (A).
