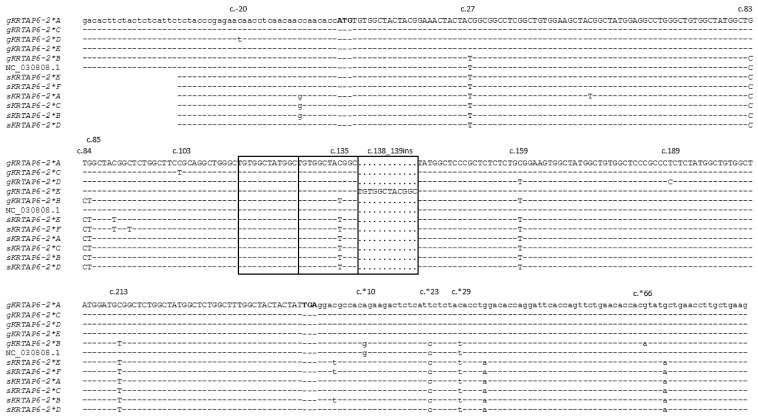
Figure 3.
Sequence alignment of the goat KRTAP6-2 variants identified in this study, together with the goat genome assembly sequence (NC_030808.1) and previously reported sheep variants. The goat sequences exclude the sequences of the primer binding regions and are labeled with the prefix ‘g’, whereas the sheep KRTAP6-2 sequences are labeled with the prefix ‘s’. Nucleotides in the coding region are represent in upper case, whereas those in the non-coding region are in lower case. Nucleotide sequences identical to the top sequence are shown by dashes and dots represent the absence of nucleotides. The locations of the nucleotide sequence differences identified among the five caprine variants are indicated above the sequences. The ‘TGTGGCTA(T/C)GGC’ repeats are shown in boxes. The transcription start codon (ATG) and stop codon (TGA) are shown in bold. The sequences of ovine variants (A–F) are retrieved from GenBank with accession numbers KT725827–KT725832.