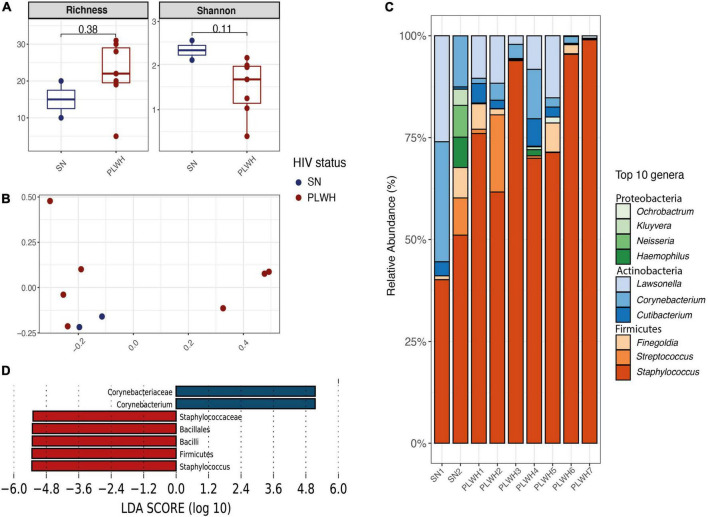
FIGURE 4.
Diversity and bacterial communities of the nasopharynx of PLWH and SN. (A) Boxplots showing the median and interquartile range. Two alpha diversity metrics were estimated: richness (number of observed species) and shannon. Groups were compared using the Wilcoxon Rank Sum test. There were no differences between PLWH and SN. (B) Principal coordinate analysis of Bray-Curtis dissimilarity, constructed from the ASV table. Samples were clustered according to their HIV status. Samples did not cluster by HIV status (PERMANOVA p = 0.407). (C) Taxa barplots showing the taxonomic composition at genus level. Only the top 10 genera are represented, these account for 99.5% of all genera present. Genera are listed from the less abundant to the more abundant within each phylum, and ordered in ascending order (Proteobacteria, Actinobacteria, and Firmicutes). Color bars reflect the phyla they belong to and the intensity of the color their relative abundance, with light colors depicting the least abundant, and the darker colors the more abundant. (D) Histogram of the LDA scores revealed two differential features taxa by HIV status: Corynebacterium was found enriched in SN while Staphylococcus enriched in PLWH. LEfSe analyses were performed with LDA values of 4.0 or higher. HIV, human immunodeficiency virus; LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; PCoA, principal coordinate analysis; PERMANOVA, permutational multivariate analysis of variance; PLWH, people living with HIV; and SN, seronegative.