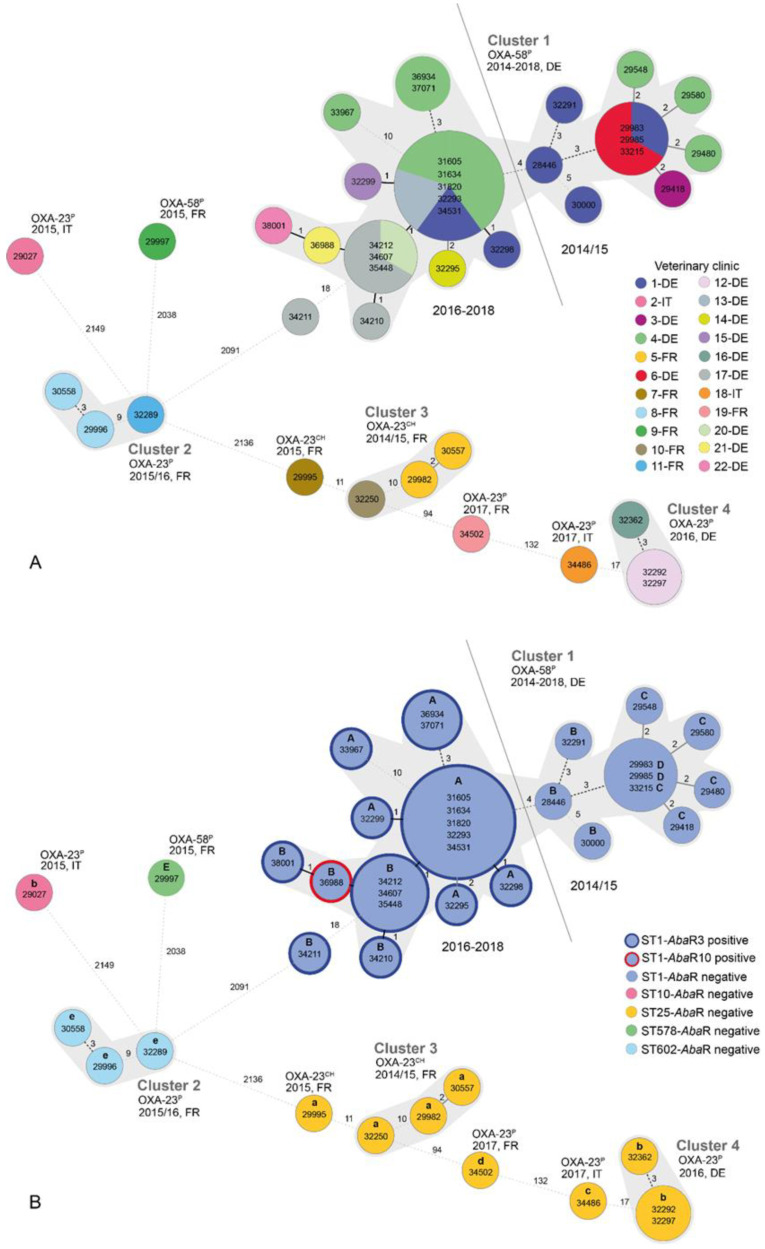
Figure 1.
Minimum spanning tree based on cgMLST allelic profiles of 42 CP-producing A. baumannii isolates. (A) Distribution of cgMLST clusters with respect to the source of isolates regarding veterinary clinic and country (Germany = DE, France = F, and Italy = IT); (B) multilocus sequence type and presence of A. baumannii resistance islands. Each circle represents an allelic profile, i.e., complex type (CT), based on sequence analysis of 2390 target genes (pairwise ignoring missing values). The numbers inside the circles represent isolate designation. The numbers on the connecting lines illustrate the number of target genes with different alleles. Clonally related CTs (≤10 allele differences) are grey shaded and numbers of clusters are indicated. The presence of CPs OXA-23 and OXA-58 and their location (P = plasmid; CH = chromosomal) as well as the year of strain isolation are indicated. Patterns of flanking regions of the blaOXA-58 (A–D) and blaOXA-23 regions (a–e) are given above the strain names in part B of the figure.