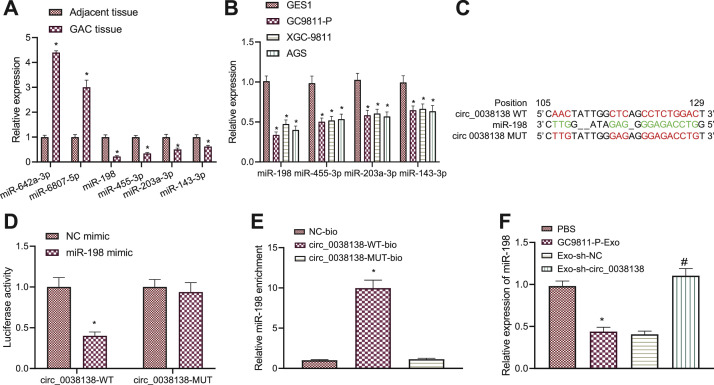
Fig. 3.
circ_0038138 competitively binds to miR-198 and suppresses its expression. A, Expression of miR-642a-3p, miR-6807-5p, miR-198, miR-455-3p, miR-203a-3p, and miR-143-3p in GAC tissues and adjacent non-neoplastic tissues determined by RT-qPCR. * p < 0.05, compared with adjacent non-neoplastic tissues. B, Expression of miR-642a-3p, miR-6807-5p, miR-198, miR-455-3p, miR-203a-3p, and miR-143-3p in GAC cells (GC9811-P, XGC-9811, AGS), and normal gastric mucosal cells (GES1) determined by RT-qPCR. * p < 0.05, compared with GES1 cells. C, The predicted binding sites or mutation sites between circ_0038138 and miR-198 from the RNA22 database. Red indicates binding sites or mutation sites. D, Binding of miR-198 to circ_0038138 confirmed by dual-luciferase reporter assay in HEK-293T cells. * p < 0.05, compared with HEK-293T cells transfected with NC mimic. E, Relative miR-198 enrichment in cells transfected with circ_0038138-WT-bio or circ_0038138-MUT-bio assessed by RNA pull-down assay. * p < 0.05, compared with cells transfected with NC-bio or circ_0038138-MUT-bio. F, Expression of miR-198 in AGS cells co-cultured with GC9811-P-Exos or Exo-sh-circ_0038138 determined by RT-qPCR. * p < 0.05, compared with AGS cells treated with PBS. # p < 0.05, compared with AGS cells co-cultured with Exo-sh-NC. The cell experiment was conducted three times independently.