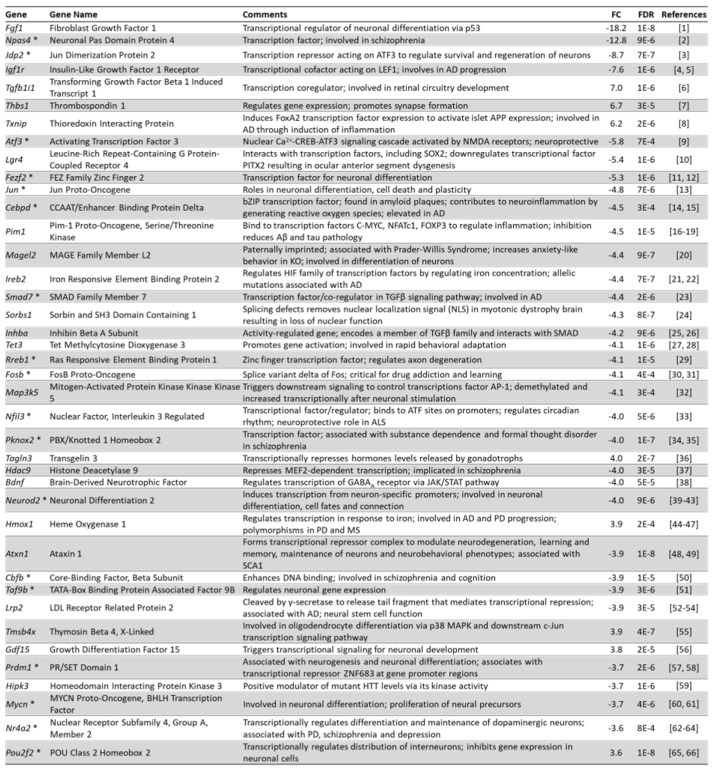
Figure 15.
Arc controls genes involved in transcriptional regulation. Shown are the top 40 genes with neuronal relevance. An asterisk (*) indicates transcription factors. ATF3: activating transcription factor 3; LEF1: lymphoid enhancer binding factor 1; FoxA2: forkhead box A2; APP: amyloid precursor protein; CREB: cAMP response element-binding protein; SOX2: SRY-box 2; PITX2: paired like homeodomain 2; bZIP: basic leucine zipper domain; C-MYC: MYC proto-oncogene, BHLH transcription factor; NFATc1: nuclear factor of activated T cells 1; FOXP3: forkhead box P3; HIF: hypoxia inducible factor; TGFβ: transforming growth factor beta; PD: Parkinson’s disease; NLS: nuclear localisation signal; SMAD: transcription factors forming the core of the TGFβ signalling pathway; AP-1: activator protein 1; ALS: amyotrophic lateral sclerosis; MEF2: monocyte enhancer factor; GABAA: γ-aminobu- tyric acid type A; JAK/STAT: Janus kinases/ signal transducer and activator of transcription; SCA1: spinocerebellar ataxia type 1; MAPK: mitogen-activated protein kinase; ZNF683: zinc finger protein 683; HTT: huntingtin. References: 1 [292], 2 [293], 3 [294], 4–5 [114,295], 6 [132], 7 [296], 8 [297], 9 [298], 10 [299], 11–12 [300,301], 13 [302], 14–15 [303,304], 16–19 [305,306,307,308], 20 [309], 21–22 [310,311], 23 [312], 24 [313], 25–26 [314,315], 27–28 [316,317], 29 [318], 30–31 [319,320], 32 [321], 33 [322], 34–35 [323,324], 36 [325], 37 [326], 38 [327], 39–43 [177,178,328,329,330], 44–47 [331,332,333,334], 48–49 [335,336], 50 [337], 51 [338], 52–54 [339,340,341], 55 [342], 56 [343], 57–58 [344,345], 59 [346], 60–61 [347,348], 62–64 [349,350,351], 65, 66 [352,353].